The students in the program approach Cells and Microbes from a variety of research angles. These topics include the molecular and cellular basis of immune responsiveness, molecular mechanisms of viral replication, host-pathogen interactions, and the control of viral, bacterial and parasitic infections.
 |
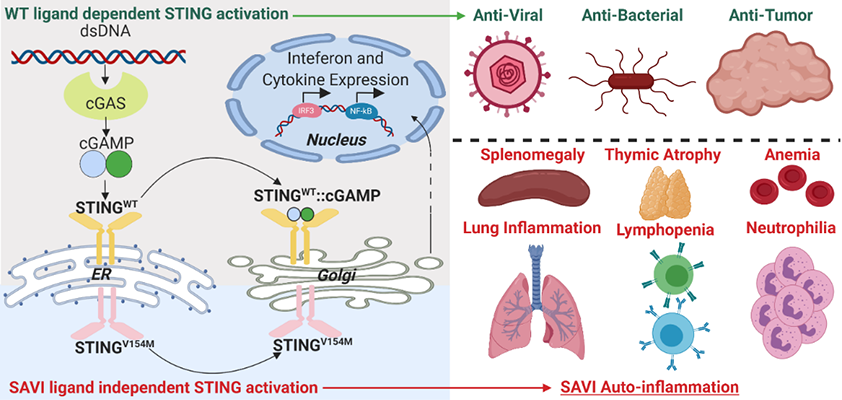 |

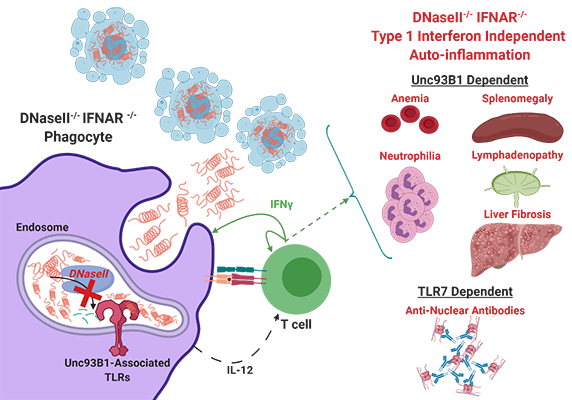
Dr. Rothstein has a long-standing interest in factors regulating the activation of autoreactive T and B cells. Her lab was the first group to demonstrate that members of the Toll-like receptor (TLR) family can detect mammalian nucleic acid ligands and that TLR9 or TLR7 play a key role in the activation of autoreactive B cells. Her current interests focus on the distinct roles of TLR7 and TLR9 in the pathogenesis of systemic autoimmune diseases, utilizing an inducible rapid onset model of cutaneous lupus. In related studies, she is exploring the role of cytosolic (eg. STING) and endosomal (TLR) sensors in models of autoinflammation resulting from constitutive activation of STING due to DNAseII deficiency or STING gain-of-function mutations. Additional projects revolve around the pro-apoptotic and pro-inflammatory functions of membrane-bound FasL and how these activities can be regulated by metalloproteinase-mediated cleavage of the extracellular FasL domain. Rothstein Lab

Kevin joined the lab in November of 2018. His thesis research is centered on the regulation of immune cells by nucleic acid (NA) pattern recognition receptors (PRRs) in the pathogenesis of auto-immune and auto-inflammatory diseases. Because microbial and host-derived NA are both abundant, organisms restrain the immune system from acting non-discriminately and have developed strategies to discern inert signals from dangerous signals. Our labs utilize two murine models which perturb these control mechanisms: STING Associated Vasculopathy with onset in Infancy (SAVI), wherein a gain-of-function mutation in STING results in constitutive activation of the cytosolic dsDNA sensing pathway cGAS-STING, and DNaseII deficiency, wherein loss of an endosomal Dnase leads to impaired NA debris clearance. Using these models, Kevin hopes to identify mechanisms by which activation of NA sensors differentially regulates adaptive and innate immune cells, and how these mechanisms converge to coordinate multi-organ inflammation.
Kevin.Gao@umassmed.edu

Dr. Fitzgerald’s has a long-standing interest in macrophages and innate immunity with a focus both on host-defense and sterile inflammation. Her group leverages cellular and molecular approaches to understand the mechanisms regulating inflammatory cytokine production. Her lab utilizes these multifaceted approaches to understand how the host responds to microbial infection and how the inflammatory process is regulated in order to avoid dangerous inflammatory responses and autoimmune disease. Her lab is particularly well-recognized for work on type I interferon gene regulation, TLR signaling pathways, inflammasome biology, long-non-coding RNAs and cytosolic DNA sensors. Ongoing projects include exploration of the molecular mechanisms that account for the autoinflammatory phenotype of mice and patients with STING gain-of-function mutations. The ultimate aim is to identify new paradigms in diagnosing and treating human disease through a deeper understanding of the molecular mechanisms regulating inflammation.
Fitzgerald Lab


We are studying the mechanisms that control autophagy, a process that is required to maintain the health of cells. Defects in autophagy lead to cell stress and have been associated with many disorders, including common and rare diseases. We use Drosophila, mammalian cell lines and mice as models to study autophagy, and our work focuses on understanding how autophagy functions in different cells and tissues, as well as under different types of cell stress.
Baehrecke Lab

Defects in autophagy, the self-degradation of cellular components, are linked to multiple disorders such as cancer, diabetes and neurodegenerative diseases. I investigate the role of vps13d, an essential gene with relatively unknown function, in context-specific autophagy in the developing Drosophila intestine. My preliminary finding suggest that this gene plays a crucial role in regulating mitophagy and mitochondrial morphology. I employ advanced Drosophila genetics, confocal microscropy and proteomics to investigate this hypothesis.
James.Shen@umassmed.edu
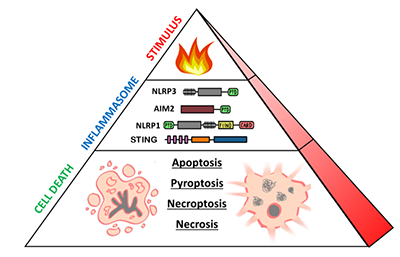

Dr. Fitzgerald is interested in the molecular mechanisms of innate immune signaling. Her lab utilizes biochemical, molecular, and genetic tools to perform research on a diversity of topics, including type I interferons, cytosolic sensors of DNA, inflammasome biology, and immune-related long-non-coding RNAs. The lab’s long-term goals are to understand how dysregulation of innate immunity contributes to the pathogenesis of infectious, autoinflammatory, and autoimmune diseases.
Fitzgerald Lab

Jeff joined the Fitzgerald lab in September 2018. His research is focused on innate immune pathways in human primary cells. In particular, he studies inflammasomes—multimeric cytosolic sensors of pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) which can elicit inflammatory cell death known as pyroptosis. He is particularly interested in the NLRP1/CARD8 proteins that are poorly conserved between murine and human systems, as well as inflammasome pathways activated by dsDNA. By pursuing human models of inflammation, Jeff hopes to bridge knowledge gaps in mouse-human immunology to improve understanding of human autoinflammatory diseases.
Jeffrey.Zhou@umassmed.edu


Dr. McCormick is a classically trained bacteriologist with a broad background and extensive experience in the study of intestinal colonization and host-microbial interactions. Her academic career has been principally committed to gastrointestinal research and she has made seminal contributions toward the understanding of how enteric bacteria (commensal, pathogenic, or probiotic) colonize the intestine and interact with the host.
Dr. McCormick’s work provided the first evidence that epithelial cells in response to pathogen contact orchestrate a pro-inflammatory program, which recruits inflammatory cells. Dr. McCormick has since identified new, previously unidentified and unexpected virulence mechanisms that are key to the inflammatory response, leading to both novel biological principles of host-microbe interactions and therapeutic intervention strategies for the treatment inflammatory bowel disease, and cancer. Her work continues to identify novel ways in which microbes interact with the intestinal epithelium, publishing over 100 original research papers and opinion pieces in this area. Dr. McCormick is an elected a Fellow of the American Academy of Microbiology, is on the Board of Editors for the journals Gastroenterology, and Gut Microbes, and serves as member of four Editorial Review Boards. She also is a permanent member of the GMPB NIH study section, and an advisor to several private foundations.
Beth McCormick Lab

My research is focused on studying patients with rare monogenic disorders causing primary immunodeficiency syndromes to understand the mechanisms and consequences of the metabolic reprogramming in immune cell activation. We identified a requirement for intact interferon gamma signaling for maximal induction of the macrophage immunometabolite, itaconate. Low levels of itaconate in patients with genetic or acquired defects in interferon gamma signaling may contribute to the infection susceptibility and immune dysregulation associated with their disease. We have also identified interferon gamma and lipopolysaccharide induced alterations in glycolytic, TCA cycle and mitochondrial metabolism in primary human monocytes and macrophages.
Katelyn.McCann@umassmed.edu
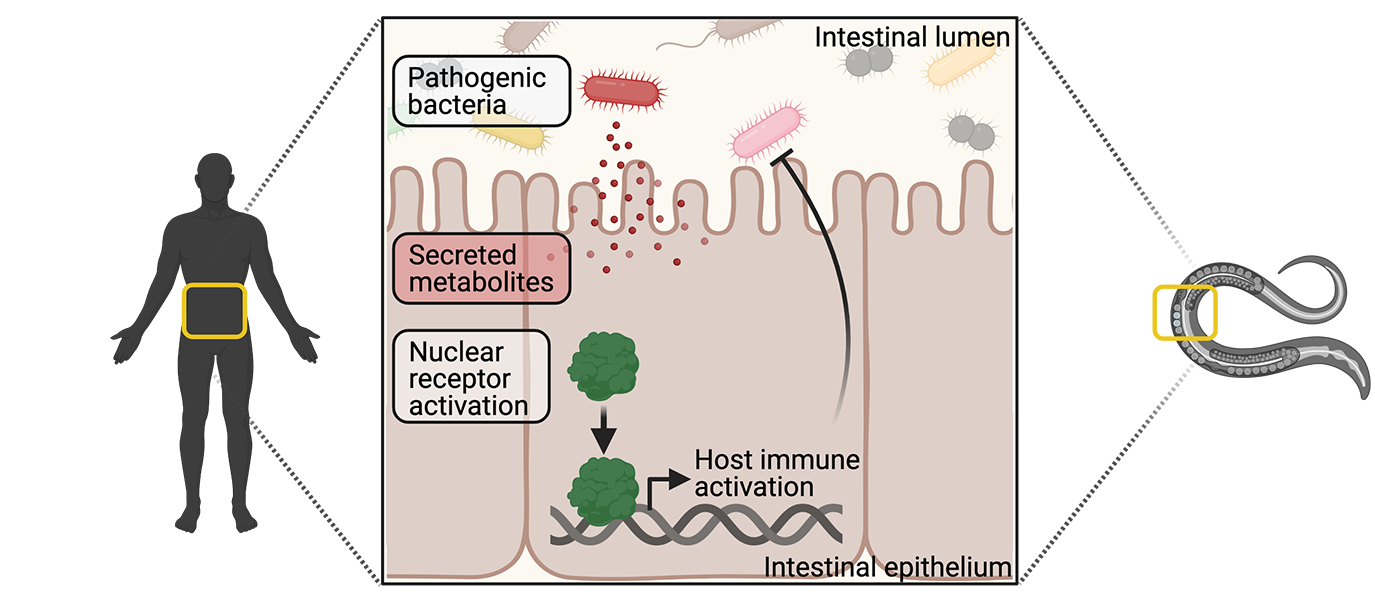

The major goal of the Pukkila-Worley laboratory is to characterize mechanisms of immune homeostasis in intestinal epithelial cells. We aim to use these discoveries to identify innate immune pathways that can be exploited to develop new therapies, which can modulate host immune activity. Our laboratory uses C. elegans as a discovery platform to define evolutionarily ancient mechanisms of pathogen sensing and immune activation.
Pukkila-Worley Lab

Intestinal epithelial cells interact with intestinal bacteria and target protective inflammatory responses towards pathogenic organisms. Interestingly, both commensal and pathogenic microbes express the so-called pathogen-associated molecular patterns (PAMPs) that are recognized by host pattern recognition receptors; however, commensal bacteria do not induce host inflammatory processes. Therefore, other mechanisms must exist that allow intestinal cells to target inflammatory defenses towards bona fide pathogens during an infection, and not harmless commensal bacteria. Here, we propose that the intestinal epithelium specifically monitors for the presence of pathogen-encoded metabolites, or their effects, to determine when the host is under attack.
Bacterial virulence determinants of particular interest are phenazine metabolites, which are produced by the pathogenic organism Pseudomonas aeruginosa. Although P. aeruginosa is the prototypical pathogen for C. elegans, the mechanism by which this nematode detects the pathogen still remains unknown. Interestingly, C. elegans encodes a greatly expanded family of nuclear receptors. In the Pukkila-Worley lab, we believe that this expansion was driven by nuclear receptors' critical roles in pathogen detection. Thus, the goal of this project is to determine the role of nuclear receptors in the detection of phenazine metabolites and activation of downstream immune responses.
Samantha.Tse@umassmed.edu