The students in the Molecular Principles Pathway are part of research projects spanning several disciplines including molecular, cellular and regulatory biochemistry; molecular biophysics; chemical biology; and structural biology. Students collaborate with several departments to research molecular pathways associated with a variety of diseases and engineer molecular systems for therapeutic use.
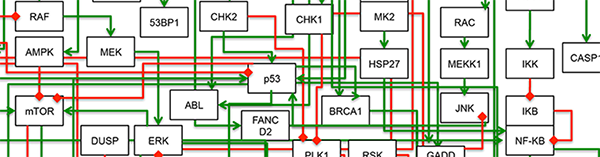

One of the promises in this post-genomic era of biology was a more personalized form of medicine, in which therapies could be tailored to the specific needs of the group (or even the individual). While a wealth of information has indeed been gathered for many types of cancer, a major shortcoming remains our inability to make rational predictions of combinatorial drug effects. Our lab is mainly interested in understanding mechanisms of anti-cancer drug action, in order to aid in the creation of better therapeutic options of patients. We focus on signaling pathways controlling the growth, survival, and death of cancer cells, in order to identify sources of therapeutic variability and to clarify the “rules” that underlie the efficacy of drugs, both when used as single agents and when used in complex combinations.
Michael J. Lee Lab

Peter is of Puerto Rican and Cuban heritage, raised in San Juan, PR and Miami, FL. He graduated from Duke University in 2011 and, subsequently, trained at The Broad Institute, where he developed his interest in cancer biology, genomics, and drug resistance. Peter joined Dr. Lee’s lab in 2015. In the lab, he is interested in understanding non-genetic mechanisms of drug resistance to targeted therapies. His thesis project centers around describing how drugs can inhibit their purported target, while simultaneously priming cells into a plastic state that allows them to adapt to the drug, leading to resistance. Peter is an active member of student government groups, leading faculty-student collaborative initiatives that advocate for students across the three schools at UMassMed. Outside of the lab, Peter enjoys watching movies, playing golf, rock climbing, and cooking.
Peter.Cruz-Gordillo@umassmed.edu

Craig Peterson received his BS from the University of Washington in 1983 and his PhD from the University of California, Los Angeles in 1988. He was a Helen Hay Whitney Foundation postdoctoral fellow from 1988-1991, in the Department of Biochemistry and Biophysics at the University of California, San Francisco. In 1992, he joined the UMass Chan Medical School as a faculty member in the Program in Molecular Medicine.
Peterson Lab

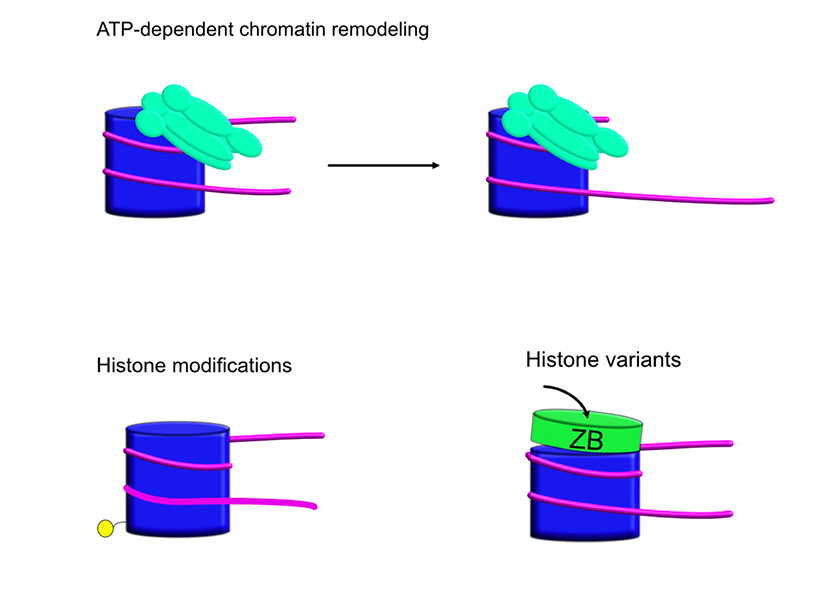
My research is broadly focused on chromatin remodeling enzymes, which are cellular machines that interact with nucleosomes in various ways. I'm also interested in how nucleosomal properties such as post-translational histone modifications or histone sequence variants affect their behavior.
Alexander.Baier@umassmed.edu

Craig Peterson received his BS from the University of Washington in 1983 and his PhD from the University of California, Los Angeles in 1988. He was a Helen Hay Whitney Foundation postdoctoral fellow from 1988-1991, in the Department of Biochemistry and Biophysics at the University of California, San Francisco. In 1992, he joined the UMass Chan Medical School as a faculty member in the Program in Molecular Medicine.
Peterson Lab

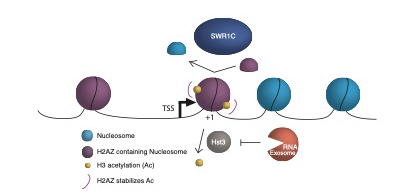
Alysia Bryll joined the Peterson lab in 2017. Her work focuses on understanding the regulatory mechanisms at promoter proximal (+1) nucleosomes that maintain transcriptional homeostasis in yeast. These nucleosomes are the site of transcriptional regulation by nucleosome modifications, the incorporation of histone variants, and post-transcriptional proteins. Her thesis work focuses on dissecting the role of the RNA exosome in regulating chromatin dynamics, specifically at these +1 nucleosomes, to maintain nascent transcript levels. Utilizing various Next generation sequencing approaches, her project will allow us to better understand these conserved mechanisms.
Alysia.Bryll@umassmed.edu

Cole received a PhD from the University of Missouri-Kansas City in 2003 and was a postdoctoral fellow in David Ron’s laboratory at the NYU School of Medicine. Cole started his own lab in the Cell Biology Program at Memorial Sloan Kettering Cancer Center in 2009. And, he joined the Department of Molecular, Cell and Cancer Biology at the UMass Chan Medical School in 2016.
He has received multiple awards for is work including the 2016 ASBMB Young Investigator, 62nd Mallinckrodt Scholar, Ellison Medical Foundation Scholar, the Boyer Award, and a Glenn Award for Research in Biological Mechanisms of Aging.
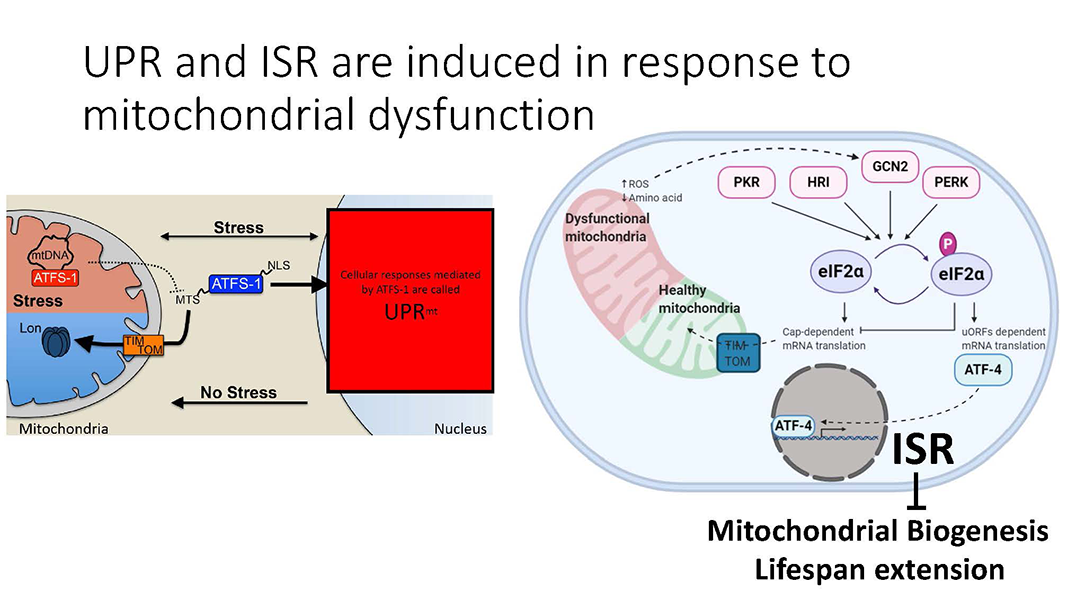
Cole Haynes Lab

Cells establish mitochondria according to their physiological needs and environment such as nutritional availabilities. Aging cells develop mitochondrial dysfunction, disabling them from adjusting to changing demands of the organism. My work involves understanding how mitochondria are built and its relation to overall lifespan of the organism.
Sookyung.Kim@umassmed.edu
We develop and apply computational methods and machine-learning techniques to analyze large-scale genomic, epigenomic, and transcriptomic data. We implement these skills to study a wide range of biological problems.
As members of the ENCODE and psychENCODE consortia, we investigate the molecular mechanisms of gene regulation by integrating and analyzing a wide variety of genome-wide deep-sequencing data including DNase-seq, ATAC-seq, CAGE, RAMPAGE, ChIP-seq, DNAme-seq, RNA-seq, eCLIP-seq, Hi-C, and single-cell sequencing data, among others. Furthermore, we study how genetic variations in the human population affect gene regulation and susceptibility for diseases. We also seek to understand the biogenesis and regulatory mechanisms of small silencing RNAs such as microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs), which we achieve by building computational pipelines to analyze diverse high-throughput sequencing datasets upon perturbing small silencing RNA pathways. We also have a long history of building algorithms and benchmarks for protein docking and design.
Zhiping Weng Lab

Thomas studies bioinformatics in Zhiping Weng’s lab. His work focuses on linking disease-associated genetic variants to disease pathology. 98% of the human genome does not code for protein, and much of the remaining genome is regulatory regions that control which genes turn on or off. Most disease-linked genetic variants do not alter protein coding genes, but rather alter non-coding regulatory regions. Thomas uses high-throughput sequencing data about gene expression, chromatin accessibility, and transcription factor binding in an effort to link disease variants with the specific cell types and pathways that are altered. As part of the PsychENCODE consortium, Thomas focuses on understanding the genetic determinants of psychiatric diseases such as autism and schizophrenia, in the hope this will lead to better treatments for these conditions.
Thomas.Reimonn@umassmed.edu