Maehr Lab Thymus Development Study Featured as Cover Story in Immunity
 Thymus development research in the laboratory of René Maehr, PhD, was featured on the cover of Immunity. The article titled A Single-Cell Transcriptomic Atlas of Thymus Organogenesis Resolves Cell Types and Developmental Maturation, highlights their cutting-edge work.
Thymus development research in the laboratory of René Maehr, PhD, was featured on the cover of Immunity. The article titled A Single-Cell Transcriptomic Atlas of Thymus Organogenesis Resolves Cell Types and Developmental Maturation, highlights their cutting-edge work.
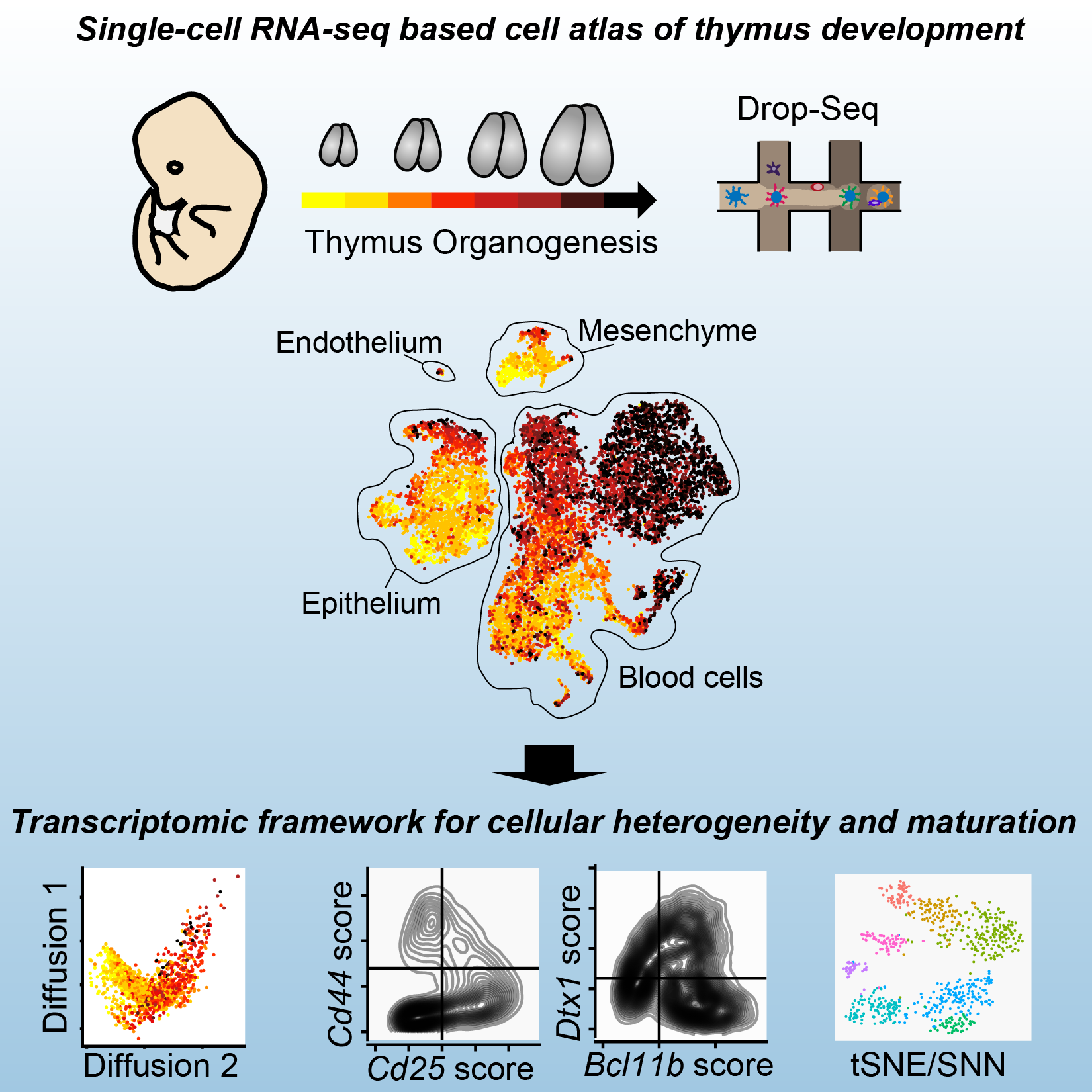
The thymus is responsible for the education and maturation of T cells, and thymus dysfunction or improper development can lead to immune disorders. Molecular details of thymus formation and maturation have been largely elusive. The Maehr lab shares a droplet-based single-cell RNA sequencing survey of mouse thymus organogenesis, covering 8 days of development from initial thymus formation to birth. In this survey, major thymic cell types were captured, including lymphocytes and epithelial, mesenchymal, and endothelial cells. The study provides a molecular framework for thymus organogenesis, offering insights into the maturation and heterogeneity of the cell types involved.
Image credit: Patrick Lane, ScEYEnce Studios
The cover illustration depicts major cellular subtypes, as captured by t-SNE analysis, as “landmasses” within an atlas, and highlights this study as a resource to help navigate the waters of thymus organogenesis.
Research Summary
Thymus development is crucial to the adaptive immune system; however, a comprehensive transcriptional framework that captures thymus organogenesis at single-cell resolution is still lacking. This cutting-edge research in the Maehr lab provides a single-cell transcriptional framework for biological discovery and molecular analysis of thymus organogenesis. Single-cell RNA sequencing (RNA-seq) was applied to capture 8 days of thymus development, perturbations of T cell receptor rearrangement, and in vitro organ cultures, producing profiles of 24,279 cells. They resolved the transcriptional heterogeneity of developing lymphocytes, and genetic perturbation confirmed the T cell identity of both conventional and non-conventional lymphocytes. The research characterized the maturation dynamics of thymic epithelial cells in vivo and classified the maturation state of cells in a thymic organ culture. It revealed the intrinsic capacity of thymic epithelium to preserve transcriptional regularity despite exposure to exogenous retinoic acid. Finally, by integrating the cell atlas with human genome-wide association study (GWAS) data and autoimmune-disease-related genes, they implicated embryonic thymus-resident cells as potential contributors to autoimmune disease etiologies. This resource provides a single-cell transcriptional framework for biological discovery and molecular analysis of thymus organogenesis.
Highlights
- Single-cell RNA-seq reveals thymic cell types and developmental trajectories
- Integration with GWASs links thymus development to autoimmune disorder etiologies
- Machine learning measures organ culture maturation of lymphocytes and epithelium