Welcome to the Lawson Laboratory website.
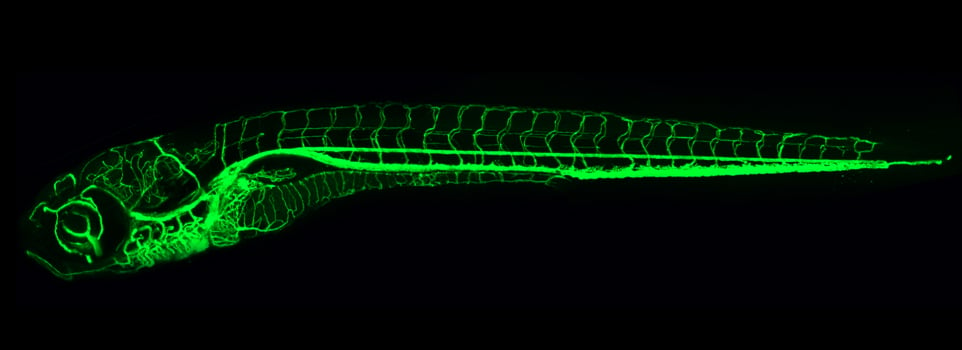
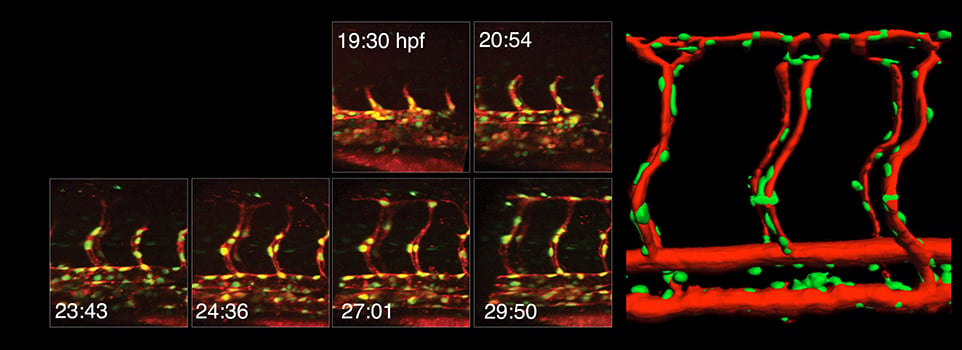
Our research focuses on vascular development using zebrafish as a model system for our studies.
Recent Publications
Shin, M., Yin, H. M., Shih, Y. H., Nozaki, T., Portman, D., Toles, B., Kolb, A., Luk, K., Isogai, S., Ishida, K., Hanasaka, T., Parsons, M. J., Burns, C. E., Burns, C. G., Wolfe, S. A., Lawson, N. D. (2023) Generation and application of endogenously floxed alleles for cell-specific knockout in zebrafish. Developmental Cell, 58(22):2614-2626.e7.
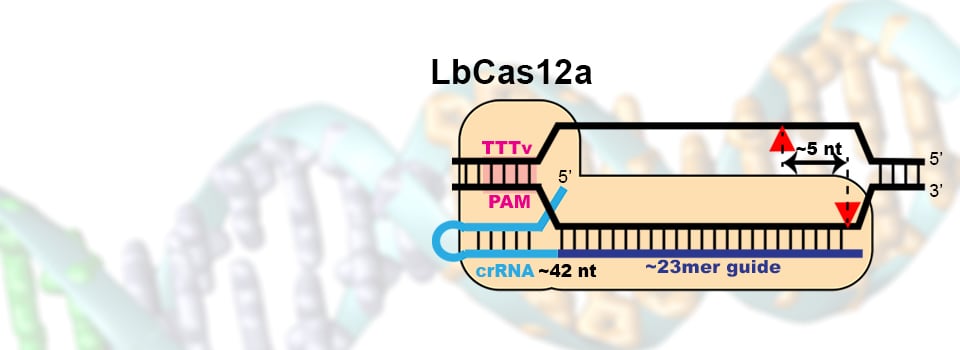
Ponnienselvan, K., Liu, P., Nyalile, T., Oikemus, S., Maitland, S. A, Lawson, N. D., Luban, J., Wolfe, S. A. (2023) Reducing the inherent auto-inhibitory interaction within the pegRNA enhances prime editing efficiency. Nucleic Acids Research, 51:6966-6980. PMC
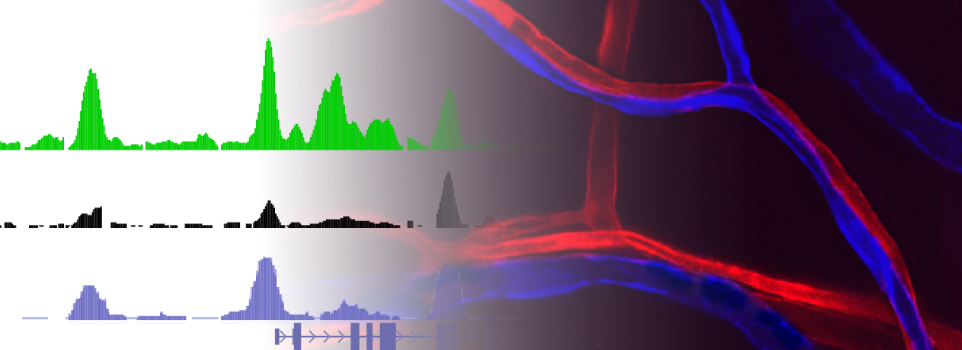
Shih, Y.-H., Portman, D., Idrizi, F., Grosse, A., Lawson, N. D. (2021) Integrated molecular analysis identifies a conserved pericyte gene signature in zebrafish. Development, 148:23:dev200189. | PMC | GEO datasets
To see all publications, click here.