Cys2His2 Zinc Finger Proteins
Cys2His2 Zinc finger proteins are the most common type of DNA-binding domain found in the majority of eukaryotic genomes. Each finger contains ~30 aa of the consensus (F/Y)-X-C-X2-5-C-X3-(F/Y)-X5-y-X2-H-X3-5-H, where X represents any amino acid and y is a hydrophobic residue, which folds into a simple beta-beta-alpha motif around a tetrahedrally coordinated single zinc ion.
The Cys2His2 zinc finger motif is a versatile scaffold for the recognition of a variety of different DNA sequences. Using the sequence of Zif268 as a framework zinc fingers with novel specificities have been created by design or by selection via phage display or bacterial two-hybrid selection from libraries in which the residues in the recognition helix have been randomized. In ?general selection, especially using a bacterial system, yields proteins with superior specificities.
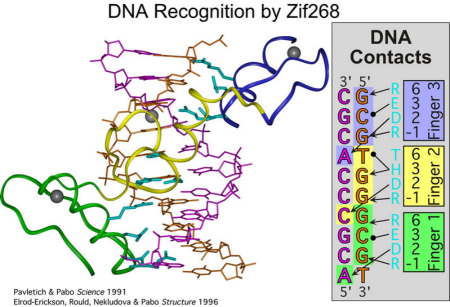
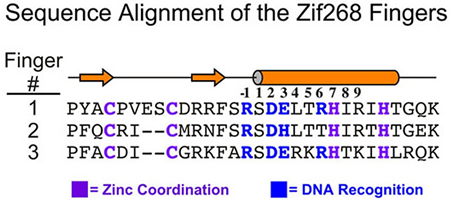
Legend for figures: The canonical zinc finger protein Zif268 recognizes its binding site using three Cys2His2 zinc fingers [DNA recognition by Zif268]. Each finger inserts its a helix into the major groove, and DNA recognition is mediated by amino acids at positions –1, 2, 3 and 6 from this helix (numbered with respect to the start of the alpha helix). The majority of contacts are made to one strand of the DNA. Interactions that are mediated by hydrogen bonds between the fingers and DNA are indicated by arrows in the panel on the right; hydrophobic interactions are indicated by open circles. An alignment of the amino acid sequence of the three Cys2His2 zinc fingers from Zif268 is shown for reference. Adjacent fingers are connected by ‘canonical’ linkers (TGQKP and TGEKP). The residues from each finger involved in defining DNA specificity are indicated in blue; residues involved in zinc coordination are indicated in purple. The secondary structure elements are indicated above the sequence with arrows indicating beta- strands and the cylinder denoting alpha-helices.