Resources
- A Resource for C. elegans Transcription Factors
- Wormflux: Metabolic Network Modeling of C. elegans
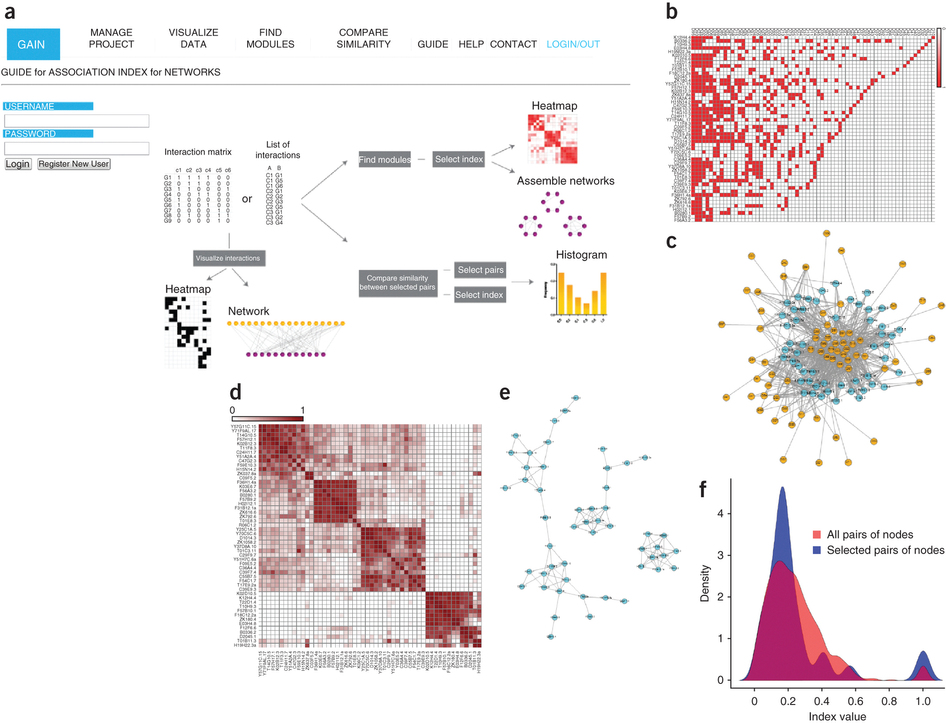
- GAIN: Guide for Association Index for Networks
- Handbook of Systems Biology
- SpotOn: an automated colony-quantification program
- MyBrid: Visualizing and managing enhanced yeast hybrid screens
- Other Technical and Methods Papers
1. A Resource of C. elegans Transcription Factors
Our most current analyses predict that the C. elegans genome encodes 941 TFs (Fuxman Bass et al., 2016). Since our initial predictions (see below), the overall number of total TFs has changed very little, illustrating the high level of gene annotation in the C. elegans genome. (Worm Transcription Factors version X.X = wTF X.X, Excel Format)
 wTF 3.0 941 TFs (Fuxman Bass et al., 2016)
wTF 3.0 941 TFs (Fuxman Bass et al., 2016)
wTF 2.2 937 TFs (Reece-Hoyes et al., 2011)
wTF 2.1 940 TFs (Vermeirssen et al., 2007)
wTF 2.0 934 TFs (Reece-Hoyes et al., 2005)
wTF 1.0 387-652 TFs (Various Sources, 1998-2000)
2. WormFlux: Metabolic Network Modeling of C. elegans
iCEL1314 is a reconstructed metabolic network model for the organism Caenorhabditis elegans. This network model contains 1,314 genes, 634 enzymes, and 2,230 metabolic reactions. WormFlux provides a web-based searchable database of iCEL1314 with detailed descriptions of model elements and their annotations in gene, enzyme, reaction, compound, and pathway pages. To facilitate applications that require modifications in biomass, we provide a ‘Biomass’ tool as part of WormFlux, which can take user-defined biomass parameters and adjusts bacterial degradation and worm biomass assembly pathways accordingly. Flux Balance Analysis, Flux Variable Analysis, and integrated gene and protein expression data will eventually be added to WormFlux. Click the figure below to access WormFlux.
Yilmaz LS, Walhout AJM. (2016). A Caenorhabditis elegans Genome-Scale Metabolic Network Model. Cell Systems 2, 297–311.
3. GAIN: Guide for Association Index for Networks
GAIN is a web tool for calculating and comparing interaction-profile similarities and defining modules of genes with similar profiles.
Fuxman Bass JI, Diallo A, Nelson J, Soto JM, Myers CL, Walhout AJ. (2013) Using networks to measure similarity between genes: association index selection. Nat Methods. 10:1169–1176. Erratum in: Nat. Methods. 11:349.
4. Handbook of Systems Biology
 Edited by Marian Walhout, Job Dekker and Marc Vidal. (2012)
Edited by Marian Walhout, Job Dekker and Marc Vidal. (2012)
Top-level scientists from different disciplines contributed 27 chapters on all aspects of systems biology. From defining systems components and their network interactions, to highly quantitative mathematical systems modeling. From yeast to plants and worms, and from cells to organisms to populations. Purchase on Amazon.
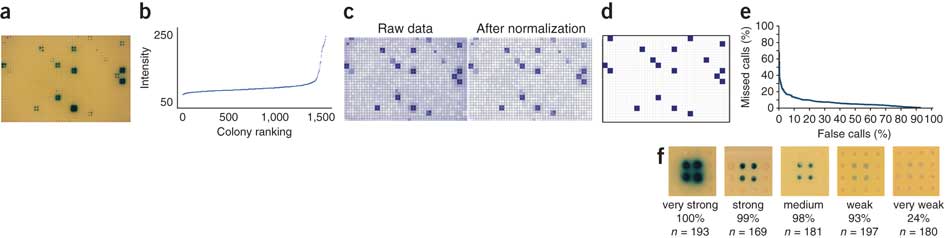
5. SpotOn
SpotOn is an automated colony-quantification program that enables the automated retrieval of positive interactions from images of enhanced yeast one-hybrid assays.
We developed a custom Perl-based program called SpotOn that automatically quantifies eY1H assay results. SpotOn imports a JPEG image of an eY1H readout plate (Fig. 4a) and performs the following tasks (See also MyBrid below). First, a grid is fitted to the 1,536 colonies to associate each colony with the transcription factor it expresses. Second, the intensity of β-galactosidase expression (that is, the blueness) of each colony is determined (Fig. 4b). Third, these intensity values are normalized for noise owing to growth differences resulting from uneven nutrient availability within each plate, as well as intrinsic differences between baits (Fig. 4c). Then, SpotOn uses a Z-score cutoff to identify positive colonies and removes false positives arising from bleed-over of blue compound from neighboring very strong positives. Finally, it identifies transcription factors for which two or more colonies score positively to produce a list of eY1H interactions (Fig. 4d).
Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, Walhout AJ. (2011) Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat. Methods. 8:1059-64.
6. MyBrid: Visualizing and managing enhanced yeast hybrid screens
MyBrid was developed for several tasks linked to these enhanced yeast hybrid assays: 1) to upload and view assay images that have been processed by SpotOn; 2) to correct (rare) miscalls by SpotOn; and 3) to download interaction datasets.
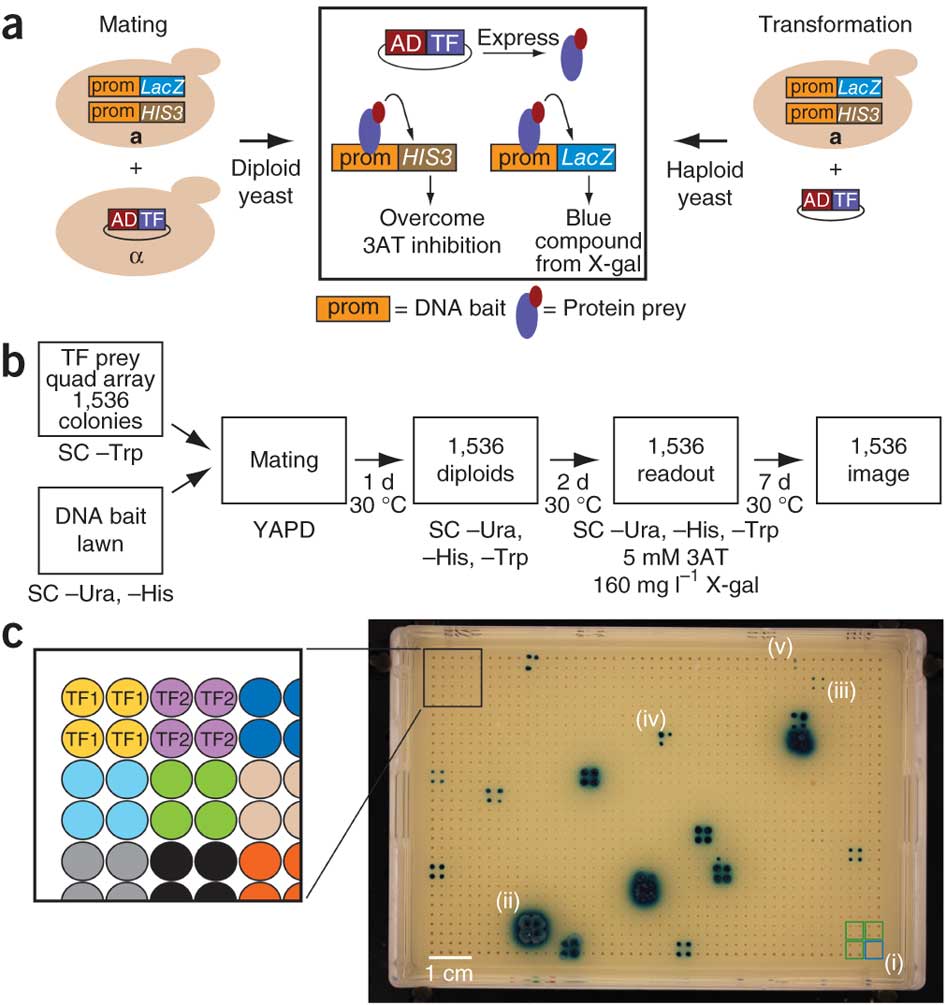
(a) Schematic of Y1H assays by mating and by transformation. prom, promoter (or DNA bait); AD, Gal4 transcription activation domain; TF, transcription factor. (b) Schematic of steps in the eY1H pipeline. SC, synthetic complete medium; –Trp, without tryptophan; –Ura, without uracil; –His, without histidine; YAPD, rich yeast medium with adenine. (c) Example of eY1H readout plate (‘1,536 image’ in b). Each transcription factor was tested in quadruplicate. (i) Control quads that lack yeast indicate plate identity and orientation (blue square), whereas yeast that contain an empty AD plasmid serve as a background above which interacting factors are detected (green squares); (ii) strong eY1H positive in which all four spots of a TF quad score positively and had bleed-over; (iii) weak eY1H positive in which all four spots of a TF quad score positively; (iv) medium eY1H positive in which three spots of a TF quad score positively; (v) very weak eY1H positive in which two spots of a TF quad score positively. We considered only quads in which at least two of the four colonies are positive because such an interaction is by definition retested.
Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, Walhout AJ. (2011) Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat. Methods. 8:1059-64.
7. Other Technical and Methods Papers
Fuxman Bass JI, Reece-Hoyes JS, Walhout AJ. (2016). Gene-Centered Yeast One-Hybrid Assay. Cold Spring Harb Protoc. 2016(12):pdb.top077669.
Conte D, MacNeil LT, Walhout AJ, Mello CC. (2015). RNA Interference in Caenorhabditis elegans. Curr. Protoc. Mol. Biol. 109:26.3.1-26.3.30.
Tamburino AM, Ryder SP, Walhout AJ. (2013). A Compendium of Caenorhabditis elegans RNA binding proteins predicts extensive regulation at multiple levels. G3. 3:297-304.
Reece-Hoyes JS, Walhout AJ. (2012). Gene-centered yeast one-hybrid assays. Methods Mol. Biol. 812:189-208.
Reece-Hoyes JS, Barutcu AR, McCord RP, Jeong JS, Jiang L, Macwilliams A, Yang X, Salehi-Ashtiani K, Hill DE, Blackshaw S, Zhu H, Dekker J, Walhout AJ. (2011). Yeast one-hybrid assays for gene-centered human gene regulatory network mapping. Nat. Methods. 8:1050-52.
Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, Liu Z, Breton G, Pruneda-Paz JL, Kim D, Kay SA, Walhout AJ, Ware D, Brady SM. (2011). Enhanced Y1H assays for Arabidopsis. Nat. Methods. 8:1053-5.
Vermeirssen V, Deplancke B, Barrasa MI, Reece-Hoyes JS, Arda HE, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Brent MR, Walhout AJ. (2007). Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat. Methods. 4:659-64.
Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJ. (2006). Gateway-compatible yeast one-hybrid screens. Cold Spring Harb. Protoc. 2006(5):pdb.prot4590.