Research
The Structure, Function and Evolution of Biological Networks
Every cell must coordinately regulate the expression and function of thousands of proteins in a robust manner to ensure cellular function. Accurate gene regulation is essential for most biological processes, including development, physiological homeostasis, and response to environmental stresses. Indeed, mis-regulated gene expression is a hallmark of many diseases, including diabetes, obesity and cancer. Proteins do not function in isolation. Instead they are wired together with other biomolecules into different types of biological networks. These networks have evolved to coordinate and integrate biological processes within a cell to ensure the function of the organism.
In the Walhout lab we aim to understand how biological networks are organized, how their organization enables function, and how these networks evolve. Our main focus is on the gene regulatory networks that control proper gene expression, and on metabolic networks that provide building blocks and energy to support organism development, growth, wound healing, homeostasis, and response to nutritional, environmental and therapeutic inputs. We are particularly interested in the communication between metabolic and gene regulatory networks to ask important questions that navigate both broad systems-level as well as deep mechanistic biological processes.
We use a variety of experimental and computational systems biology approaches to dissect complex biological networks. These include large-scale genetic screens, metabolomics, and transcription factor binding assays, as well as flux balance analysis, nested effects modeling, and statistics.
We mainly use the roundworm Caenorhabditis elegans as a model system. Worms are highly adaptable, easy to manipulate, and have many analogs in human genetics. Furthermore, there are many genetic tools and worm-specific techniques that are not available for studying “higher” eukaryotes. Overall, our research involves three broad areas in systems biology:
1: NETWORK STRUCTURE, FUNCTION AND EVOLUTION
2: TISSUE-RELEVANT NETWORKS
3: HOST-MICROBIOTA INTERACTIONS IN NUTRITION AND DRUG RESPONSE
1: NETWORK STRUCTURE, FUNCTION AND EVOLUTION
The structure of networks is key to their function. For instance, specific network subgraphs have been found to occur over and over again in different biological networks, likely because their functionality has proved to provide benefit to the organism over evolutionary time. We study different aspects of network structure or organization and its impact on C. elegans gene expression, metabolism and physiology. Most notably, we have discovered a persistence detector circuit comprised of a type 1, coherent feedforward loop with an AND-logic gate in which two transcription factors function to that facilitate metabolic network rewiring, only when needed (see more below, Bulcha et al., 2019).
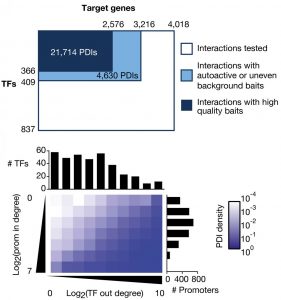
Gene Regulatory Networks
In gene regulatory networks, genes and their regulators are the nodes and the physical or functional interactions between them are the edges. These networks are bipartite and directional. We have pioneered the use of gene-centered yeast one-hybrid assays to identify physical interactions between transcription factors and regulatory regions in the genome such as promoters and introns. These assays start, for instance, with a promoter of interest and identify the collection of transcription factors with which this promoter can interact. This is in contrast to more widely used protein-centered methods such as Chromatin Immunoprecipitation (ChIP) that identify for a single transcription factor the different genomic regions with which it can interact. We have delineated a large promoter-transcription factor network for C. elegans (Reece-Hoyes et al., 2013; Fuxman Bass et al., 2016). This network forms the foundation for in-depth studies about the organization and evolution of gene regulatory networks, and provides a treasure of interactions that are not yet explored in detail. A major focus in the lab has been on gene families such as microRNAs (Martinez et al., 2008a; Martinez et al., 2008b), helix-loop-helix transcription factors (Grove et al., 2009; De Masi et al., 2011) and insulins (Ritter et al., 2013). Future studies will focus on the regulation of metabolic gene families such as dehydrogenases and dehydratases.
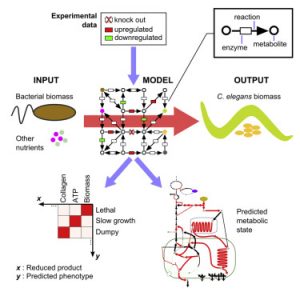
Metabolic Networks
The metabolic network is the collection of reactions that converts our diet into building blocks for growth and would healing, and energy to support cellular and organismal activities. In metabolic networks, metabolites are the nodes and the reactions between them are the edges. Genes are annotated to encode enzymes that catalyze specific metabolic reactions based on homologies with other organisms. We have reconstructed the C. elegans metabolic network and developed a searchable web tool called Wormflux with which this network can be explored (Yilmaz & Walhout, 2016). We named this model iCEL1273 because it contains 1273 genes. In addition, it contains 623 enzymes (some enzymes are multiprotein complexes) and 1985 metabolic reactions involving 887 metabolites. We predict that the C. elegans genome encodes more than 2,000 metabolic enzymes. Future studies will incorporate additional genes into the model, and test network function under different nutritional, environmental or genetic conditions.
Network Communication
Metabolism is controlled by gene expression and vice versa. Therefore, there is likely extensive communication between gene regulatory and metabolic networks, but this communication remains relatively unexplored. We have previously delineated interactions between metabolic gene promoters and transcription factors using yeast one-hybrid assays (Arda et al., 2010). This study revealed an enrichment for nuclear hormone receptor (NHR) transcription factors. C. elegans has more than 270 NHRs, while humans have 48 and flies have 18. A major interest in the lab is to understand the function of these NHRs in the context of both gene regulation and metabolism. In addition, we are interested in exploring more broadly how much of metabolism is regulated by gene expression, and how much of gene expression responds to metabolism.
Recent publications:
Bulcha JT, Giese GE, Ali MZ, Lee Y-U, Walker MD, Holdorf AD, Yilmaz LS, Brewster RC, Walhout AJM. (2019). A Persistence Detector for Metabolic Network Rewiring in an Animal. Cell Reports 26, 460-468.
Fuxman Bass, JI, Pons, C, Kozlowski, L, Reece‐Hoyes,JS, Shrestha, S, Holdorf, AD, Mori, A, Myers, CL, Walhout, AJM. (2016). A gene‐centered C. elegans protein–DNA interaction network provides a framework for functional predictions. Mol Sys Biol 12: 884.
Yilmaz LS, Walhout AJM. (2016). A Caenorhabditis elegans Genome-Scale Metabolic Network Model. Cell Systems 2, 297–31.
Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, Diprima S, Dricot A, Lajoie BR, Ribeiro PS, Weirauch MT, Hill DE, Hughes TR, Myers CL, Walhout AJ. (2013). Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell 51:116-27.
2: TISSUE-RELEVANT NETWORKS
Different tissues have different functions. From muscle enabling movement of heart and limbs to the gastrointestinal tract absorbing and delivering nutrients and liver in central metabolism and detoxification. We aim to connect network structure and organization to tissue-relevant functions.
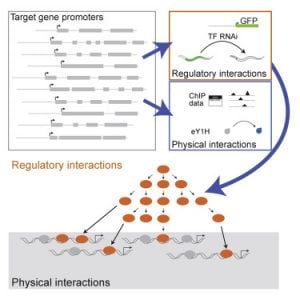
Gene Regulatory Networks
We have previously used yeast one-hybrid assays to delineate the first gene-centered, physical gene regulatory network for the C. elegans intestine (Deplancke et al., 2006). More recently, we have moved from physical interactions between intestinal gene promoters and transcription factors, to functional interactions. Again, we have pioneered the use of gene-centered methods, in this case combining transgenic worms expressing fluorescent proteins in the intestine with systematic transcription factor RNAi (MacNeil et al., 2015). This study forms the foundation for ongoing work in the lab as it really harnesses the power of C. elegans – there are no other models with which one can look at the effect of transcription factor RNAi on gene expression in a specific tissue in living animals. We learned that physical interactions between transcription factors and intestinal gene promoters do not always have a (measurable) effect on promoter activity under the conditions tested, and conversely that many transcription factors affect the activity of the promoter without binding to it. This indicates that there are layers of regulation, and we are currently further exploring this.
Metabolic Networks
Different tissues have different metabolic functions. We are currently developing metabolic network models for different C. elegans tissues to gain more insight into metabolic network compartmentalization in this animal.
Network Communication
Metabolism affects gene expression and vice versa. We are currently exploring the extent of transcriptional regulation of metabolism. In addition, we are using gene-centered approaches to investigate how the perturbation of metabolism affects C. elegans intestinal gene expression. Finally, we are systematically exploring the effects of metabolic perturbations on gene expression.
Recent publications:
MacNeil LT, Pons C, Arda HE, Giese GE, Myers CL, Walhout AJM. (2015). Transcription Factor Activity Mapping of a Tissue-Specific In Vivo Gene Regulatory Network. Cell Systems 1, 152-162.
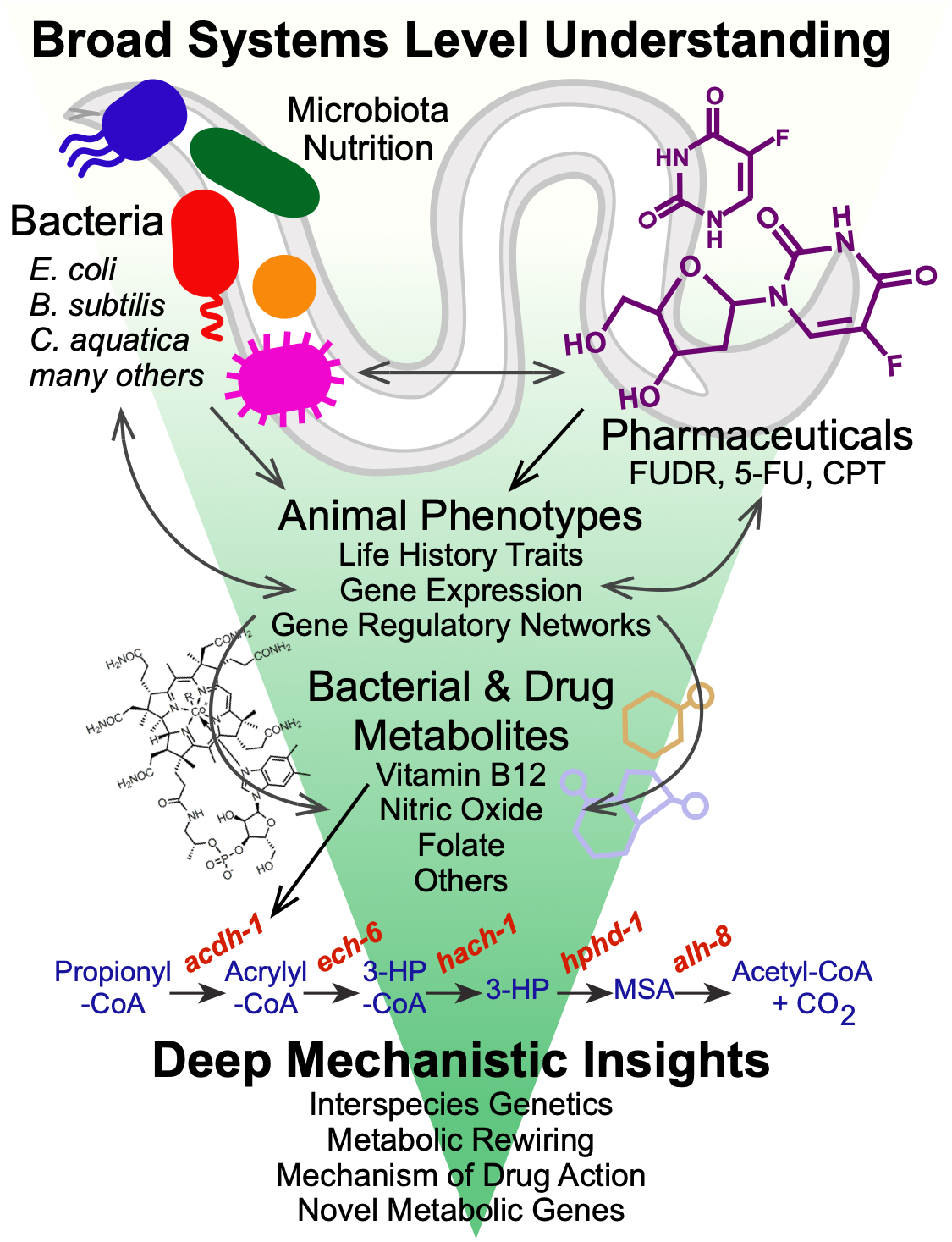
3: HOST-MICROBIOTA INTERACTIONS IN NUTRITION AND DRUG RESPONSE
Nutrition
Diet plays a fundamental role in our development, physiology and performance. The bacteria that inhabit our gut, known as our gut microbiota, play a critical role in digestion, as well as in numerous other processes including the response to different therapeutic drugs (see below). Specifically, they break down plant fibers that we cannot digest ourselves, which results in the production of short chain fatty acids such as propionate and butyrate. We use C. elegans and its bacterial diet as a model for nutrition, as well as for the interactions between bacteria and host cells, such as those occurring in the human gastrointestinal tract.
We discovered that feeding C. elegans different species of bacteria can result in major differences in life history traits, including development, reproduction, and aging (MacNeil et al., 2013). Using a combination of worm genetics (Watson et al., 2013), and bacterial genetics in both diets (Watson et al., 2014), we discovered that vitamin B12 is synthesized by only some diets, and that vitamin B12 accelerates worm development and alters metabolic gene expression.
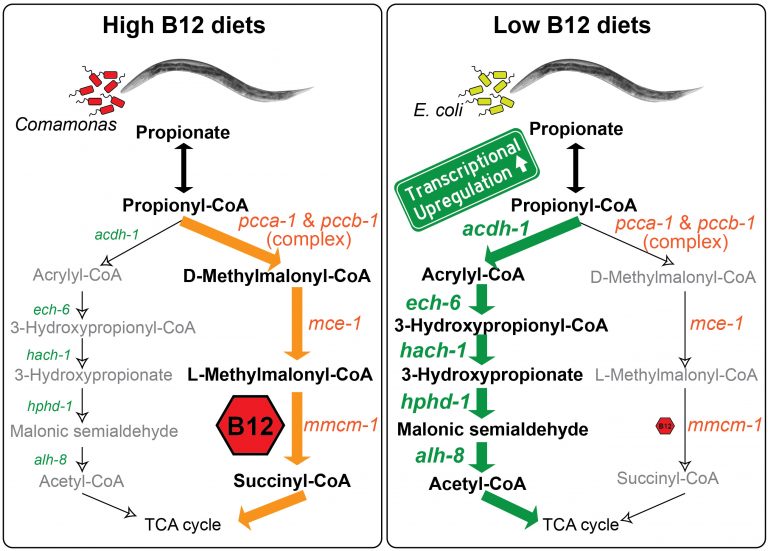
We discovered that C. elegans has two metabolic pathways to break down the short chain fatty acid propionate. Propionate buildup is a product of amino acid catabolism, and too much propionate is toxic to both worms and mammals. The canonical mechanism of propionate breakdown that we also use is dependent on vitamin B12, and patients with mutations in the enzymes in this pathway are born with devastating, often fatal, acidemia diseases. When C. elegans consumes a bacterial diet low in vitamin B12, or when the canonical pathway is genetically perturbed, it transcriptionally rewires its metabolism by activating a vitamin B12-independent propionate shunt. This gives the animal metabolic plasticity to rewire metabolism depending on available nutrients (Watson et al., 2016).
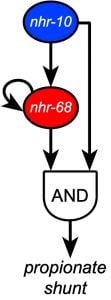
Recently, we discovered that two NHR transcription factors, nhr-10 and nhr-68, function in specific gene regulatory network circuit known as a type 1, coherent feed-forward loop with AND-logic gate to form a persistence detector that ensures that the propionate shunt is only activated when necessary (Bulcha et al., 2019).
Current studies focus on propionate independent regulation of the other vitamin B12-dependent pathway – the one carbon or methionine/SAM cycle that is critical for the generation of methyl groups used to synthesize membrane components and modify histones and other molecules. In addition, we study how other parts of the metabolic network influence metabolic gene expression and functionality with a focus on leucine and glycine catabolism. We have set up metabolomics using gas chromatography/mass spectrometry (GC-MS) to directly probe the worm metabolome.
We recently identified more than 200 E. coli mutants that slow C. elegans development when used as diet (Zhang et al., 2019). Surprisingly, we found that most of these mutants slow worm growth by providing low amounts of dietary iron, high levels of reactive oxygen species (ROS), or both. We are currently investigating the connections between bacterial metabolism and worm growth in more detail. In addition, we are interested in the interplay between ROS generated in bacteria and ROS formed in the worm itself.
Drug Response
We discovered that worms respond markedly differently to chemotherapeutic drugs when fed different bacterial diets. For instance, when we tested a panel of 11 drugs, we found that 4 affected the worms at the dose tested. Three drugs reduced fecundity and of these three, the effect of two was modulated by bacterial diets, and in opposite ways meaning one diet increased toxicity of one drug but enhanced the effect of the other. We then used bacterial genetics to unravel the underlying mechanism by which bacteria modulate the effect of these drugs and found that bacterial ribonucleotide metabolism was key (García-González et al., 2017). We are currently studying additional drugs and bacteria, and hope to expand these studies in the future.
Recent publications:
Zhang J, Li X, Olmedo M, Holdorf AD, Shang Y, Artal-Sanz M, Yilmaz LS, Walhout AJM. (2019). A Delicate Balance between Bacterial Iron and Reactive Oxygen Species Supports Optimal C. elegans Development. Cell Host Microbe 26, 400-411.
Bulcha JT, Giese GE, Ali MZ, Lee Y-U, Walker MD, Holdorf AD, Yilmaz LS, Brewster RC, Walhout AJM. (2019). A Persistence Detector for Metabolic Network Rewiring in an Animal. Cell Reports 26, 460-468.
García-González AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, Walhout AJM. (2017). Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell 169, 431-41.
Watson E, Olin-Sandoval V, Hoy MJ, Li C, Louisse T, Yao V, Mori A, Holdorf AD, Troyanskaya OG, Ralser M, Walhout AJM. (2016). Metabolic network rewiring of propionate flux compensates vitamin B12 deficiency in C. elegans. eLife 2016;5:e17670.
Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJ. (2014). Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156:759-70.
Watson E*, MacNeil LT*, Arda HE, Zhu LJ, Walhout AJ. (2013). Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell 153:253-266.
MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJ. (2013). Diet-Induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153:240-252.