Join us!
 We strive to provide a constructive, respectful, and collegial environment, empowering scholars to work together to fulfill their potential as scientists having an outstanding impact both in fundamental research and translating to medicine.
We strive to provide a constructive, respectful, and collegial environment, empowering scholars to work together to fulfill their potential as scientists having an outstanding impact both in fundamental research and translating to medicine.
What we do
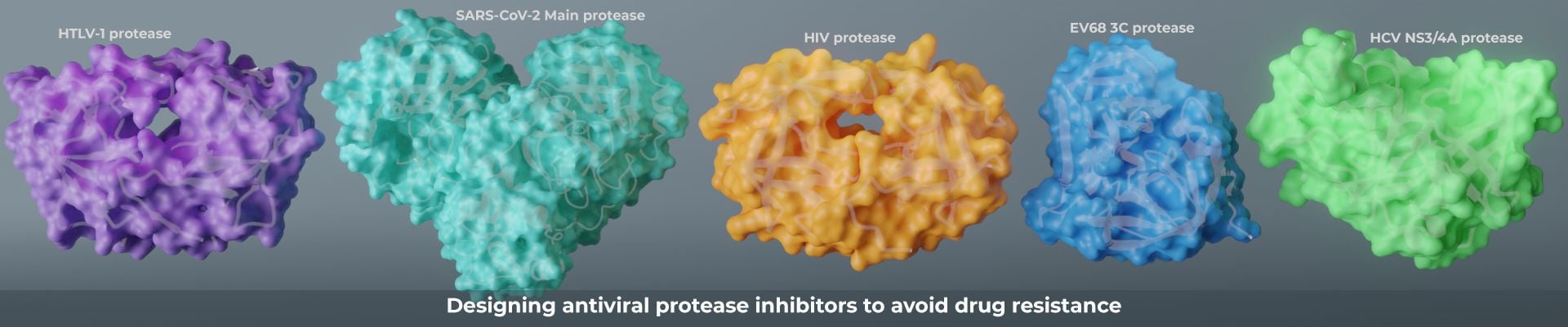
 We aim to understand the molecular basis of drug resistance and to use our new paradigm of drug design to minimize the evolution of that resistance. Resistance occurs when a heterogeneous population of a drug target is challenged by the selective pressure of a drug. In cancer and viruses this heterogeneity is partially caused APOBEC3’s. In our studies of viral proteases which include among others HIV, HCV and SARS-CoV-2. We discovered resistance mutations occur either where drugs physically contact regions of the drug target that are not essential for substrate recognition or alter the ensemble dynamics of the drug target favoring substrate. We leverage these insights into new strategies in structure-based drug design to minimize the likelihood of resistance by designing inhibitors to stay within what we define as the substrate envelope. Our new paradigm of drug design minimizes chances of resistance. Realizing that disrupting the drug target’s activity is necessary but not sufficient for developing a robust drug that avoids resistance.
We aim to understand the molecular basis of drug resistance and to use our new paradigm of drug design to minimize the evolution of that resistance. Resistance occurs when a heterogeneous population of a drug target is challenged by the selective pressure of a drug. In cancer and viruses this heterogeneity is partially caused APOBEC3’s. In our studies of viral proteases which include among others HIV, HCV and SARS-CoV-2. We discovered resistance mutations occur either where drugs physically contact regions of the drug target that are not essential for substrate recognition or alter the ensemble dynamics of the drug target favoring substrate. We leverage these insights into new strategies in structure-based drug design to minimize the likelihood of resistance by designing inhibitors to stay within what we define as the substrate envelope. Our new paradigm of drug design minimizes chances of resistance. Realizing that disrupting the drug target’s activity is necessary but not sufficient for developing a robust drug that avoids resistance.
How we do it
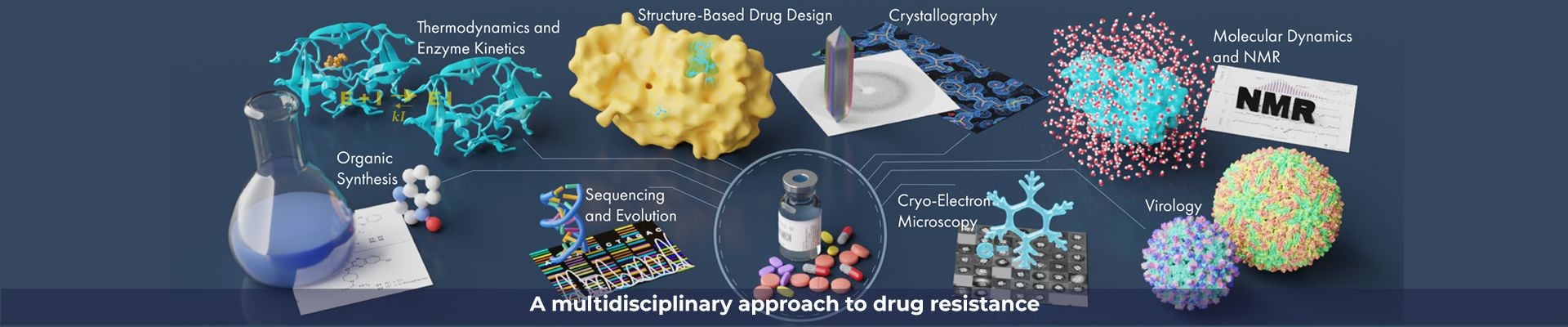
 We strive to apply a synergistic combination of various experimental and computational methods. We combine the experimental techniques of protein crystallography, cryoEM/ET, organic chemistry, enzymology, deep sequencing, and other biophysical techniques, with the computational strategies of parallel molecular dynamics simulations, homology modeling and docking, to elucidate the molecular mechanisms of drug resistance and develop new inhibition strategies.
We strive to apply a synergistic combination of various experimental and computational methods. We combine the experimental techniques of protein crystallography, cryoEM/ET, organic chemistry, enzymology, deep sequencing, and other biophysical techniques, with the computational strategies of parallel molecular dynamics simulations, homology modeling and docking, to elucidate the molecular mechanisms of drug resistance and develop new inhibition strategies.
Why we do it
 Drug resistance is a major obstacle in modern medicine, negatively impacting the lives of millions of patients and costing our society billions of dollars each year. This often happens under the selective pressure of therapy in heterogenous bacterial, viral and fungal infections and cancer due to their rapid evolution. In many cases, resistance to drugs develops so rapidly that our most valuable drugs become obsolete shortly after their introduction to clinic. Instead of considering resistance only after a drug fails, we need a paradigm shift to incorporate preemptive strategies into drug design to avoid resistance.
Drug resistance is a major obstacle in modern medicine, negatively impacting the lives of millions of patients and costing our society billions of dollars each year. This often happens under the selective pressure of therapy in heterogenous bacterial, viral and fungal infections and cancer due to their rapid evolution. In many cases, resistance to drugs develops so rapidly that our most valuable drugs become obsolete shortly after their introduction to clinic. Instead of considering resistance only after a drug fails, we need a paradigm shift to incorporate preemptive strategies into drug design to avoid resistance.