Molecular illustrations on this webpage were generated by Leonora Martínez-Núñez, PhD.

We've realized that through understanding the molecular mechanism by which the disease process occurs we could develop inhibitors that block the disease in such a way that the likelihood of resistance occurring is greatly reduced.
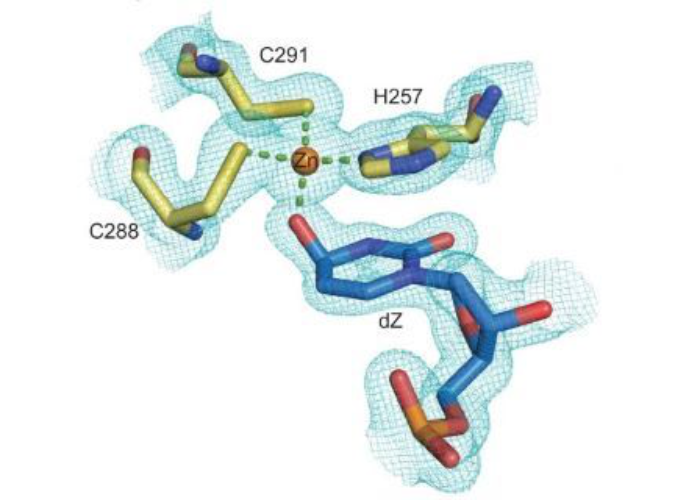
Maiti A, Hedger AK, Myint W, Balachandran V, Watts JK, Schiffer CA, Matsuo H. Nat Commun. 2022
oi: 10.1038/s41467-022-34752-1. PMID: 36402773; PMCID: PMC9675756.
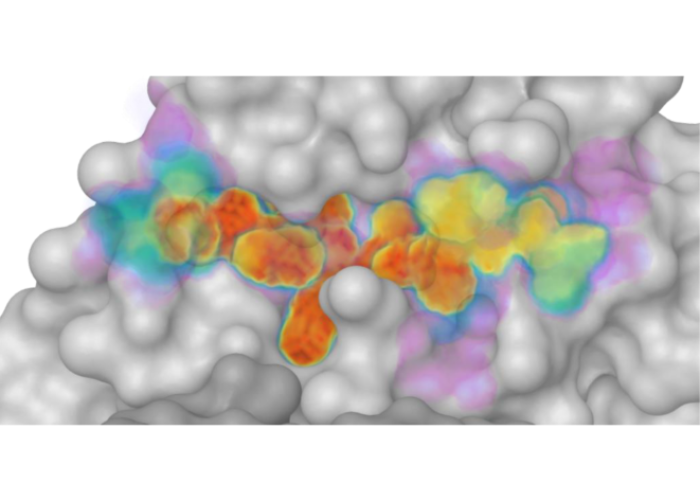
Defining the substrate envelope of SARS-CoV-2 main protease to predict and avoid drug resistance
Shaqra AM, Zvornicanin SN, Huang QYJ, Lockbaum GJ, Knapp M, Tandeske L, Bakan DT, Flynn J, Bolon DNA, Moquin S, Dovala D, Kurt Yilmaz N, Schiffer CA. Nat Commun. 2022
doi: 10.1038/s41467-022-31210-w. PMID: 35729165; PMCID: PMC9211792.
Qiuyu J. Huang, Kangkang Song, Chen Xu, Daniel N.A. Bolon, Jennifer P. Wang, Robert W. Finberg, Celia A. Schiffer, Mohan Somasundaran. Structure March 2022.
DOI: 10.1016/j.str.2022.02.014
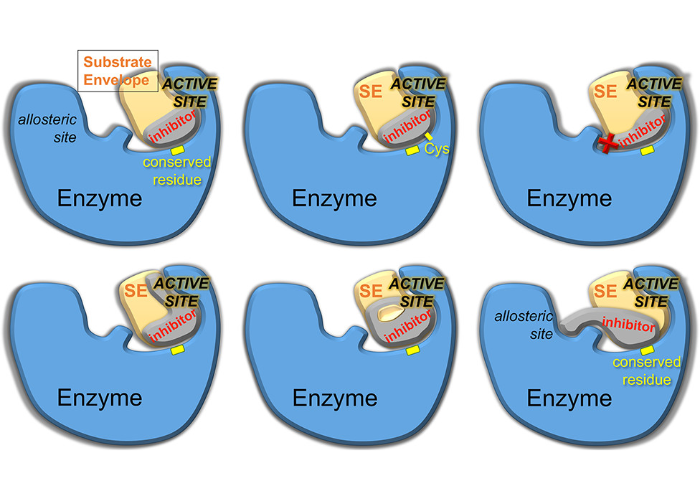 Drug Design Strategies to Avoid Resistance in Direct-Acting Antivirals and Beyond
Drug Design Strategies to Avoid Resistance in Direct-Acting Antivirals and Beyond










Molecular illustrations on this webpage were generated by Leonora Martínez-Núñez, PhD.