Introducing Sequence Diversity: APOBEC3s
Host Factors (APOBEC3s): Enzymes that Introduce Diversity into Genomes
APOBEC3s are Zn2+ dependent cytidine deaminases that are among the fastest evolving human proteins with diverse biological functions and implications for cancer and immunity. APOBEC3 (A3) cytidine deaminases are powerful DNA mutators with key roles in our immunity during antigen-driven antibody diversification as well as in innate antiviral and antibacterial defense. A3 proteins catalyze deamination of deoxy-cytidine in single-stranded DNA (ssDNA) resulting in cytidine to uridine mutation. These changes add diversity to viral and cellular genomes that likely contribute to both viral and cancer drug resistance.
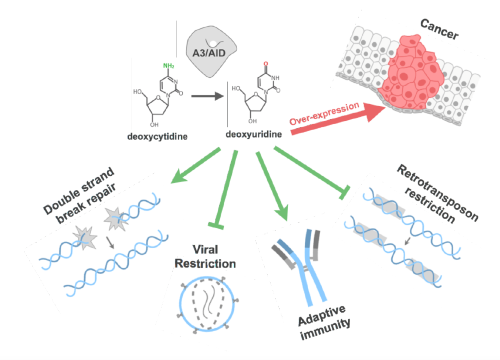
Four of the seven human A3s restrict HIV-1 by 'hypermutating' the reverse-transcribed viral genomic DNA. This introduction of viral diversity may inadvertently help the virus to evolve drug resistance more readily. HIV-1 Vif counters this host restriction factor by targeting APOBEC3s to proteasomal degradation. Two of the human A3s are implicated in cancer. However, there is no apparent correlation between biological function, catalytic activity, Vif binding, and sequence similarity between A3 domains.
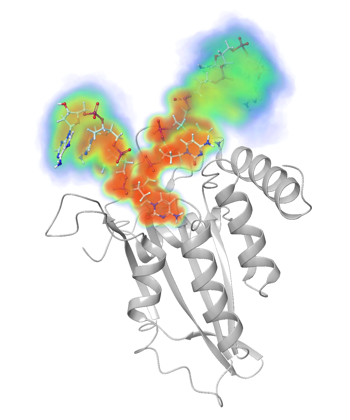 APOBEC3 bound to ssDNA |
Our research is focused on understanding the structural basis of biological function for APOBEC3 proteins. Together with our collaborator Dr. Hiroshi Matsuo (NCI) we lead the field in determining structures of APOBEC3 domains and strive to elucidate with integrated strategies how the structures of these domains influence the specificities and function of these enzymes. We solved the crystal structures of three APOBEC3 domains (APOBEC3G-CTD, APOBEC3F-CTD and APOBEC3A in apo and DNA-bound forms) and modeled the rest of the structures. These structures have gained us unique insights into the activity, specificity and oligomerization of these enzymes. In addition to structural/computational analysis and biochemical characterization, we are screening for potential selective inhibitors that may be initial therapeutic leads. |
Manuscript Highlights
 |
|
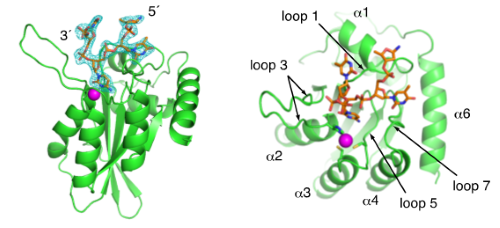
Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. |
 |
|
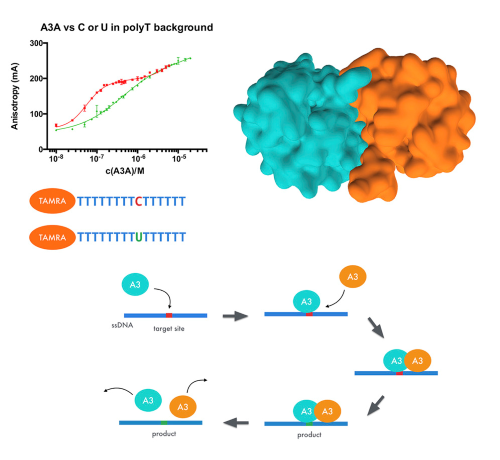
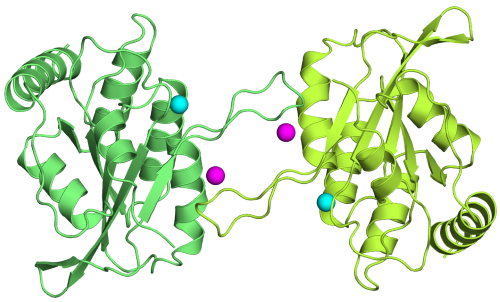
The ssDNA Mutator APOBEC3A Is Regulated by Cooperative Dimerization. |
 |
|
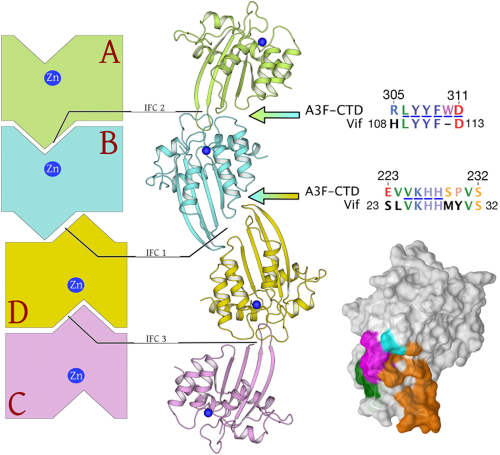
Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. |
 |
|
Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. |

