Elizabeth Hillman
Professor of Biomedical Engineering and Radiology; Principal Investigator at Columbia's Zuckerman Institute, Laboratory for Functional Optical Imaging, Columbia University
http://orion.bme.columbia.edu/~hillman/index.html
Our research focuses on capturing functional information about living tissues using optical techniques. A major theme of our lab is in-vivo neuroimaging, in particular examination of the relationship between blood flow changes in the brain and underlying neuronal activity. This work has led us to develop a range of advanced in-vivo imaging technologies including laminar optical tomography and hyperspectral two-photon microscopy. We are exploring additional applications for these technologies including clinical and pre-clinical imaging of living skin. We are also developing techniques for non-invasive 'molecular imaging' of small animals to allow improved studies of disease and development of new treatments and drugs.
Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0.
Voleti V, Patel KB, Li W, Perez Campos C, Bharadwaj S, Yu H, Ford C, Casper MJ, Yan RW, Liang W, Wen C, Kimura KD, Targoff KL, Hillman EMC. Nat Methods. 2019 Oct;16(10):1054-1062. doi: 10.1038/s41592-019-0579-4.
Bouchard MB, Voleti V, Mendes C, Lacefield C, Grueber W, Mann R, Bruno R, Hillman EMC, “Swept, confocally aligned planar excitation (SCAPE) microscopy for ultra-fast, volumetric microscopy of behaving organisms”, Nature Photonics, 9, 113–119 (2015).
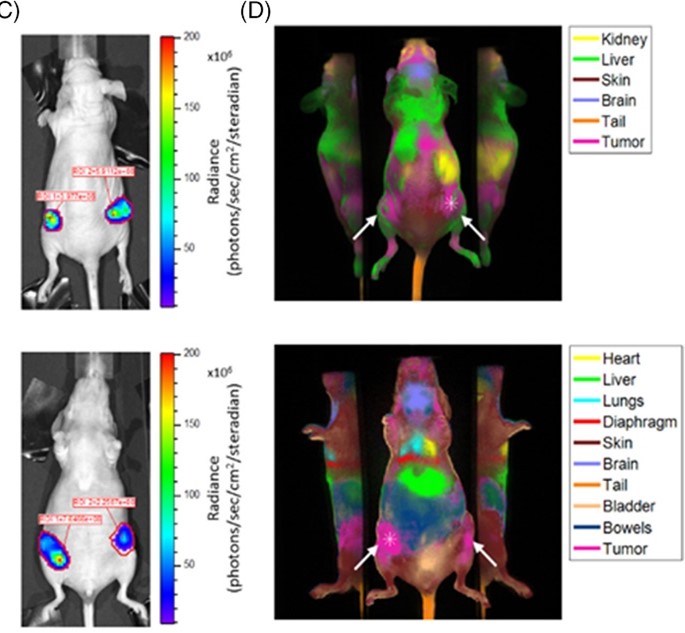
Perfusion‐based fluorescence imaging method delineates diverse organs and identifies multifocal tumors using generic near‐infrared molecular probes
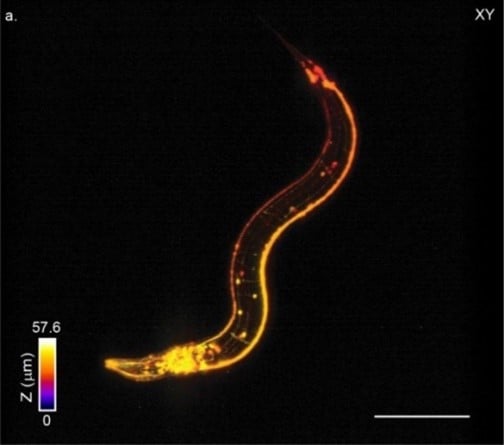
The high pixel band¬width of the HiCAM camera enabled dense sampling over a large 525x465x60 µm of view at 10 VPS. Image shows a single volume from a time-series on a freely moving C. elegans worm expressing panneuronal GFP (IM324: edls20[F25B3.3::GFP+rol-6(su1006)]). The colors represent depth into the sample and the data set has been square-rooted for visualization purposes to enhance contrast between the bright cell bodies and the dimmer dendritic processes.
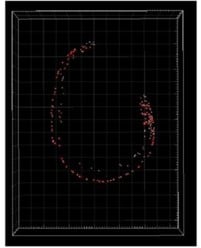
Supervised automated cell tracking over 2 minutes of dual-color SCAPE 2.0 data acquired on an unrestrained C. elegans at 26.4 VPS. From: Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0