Research
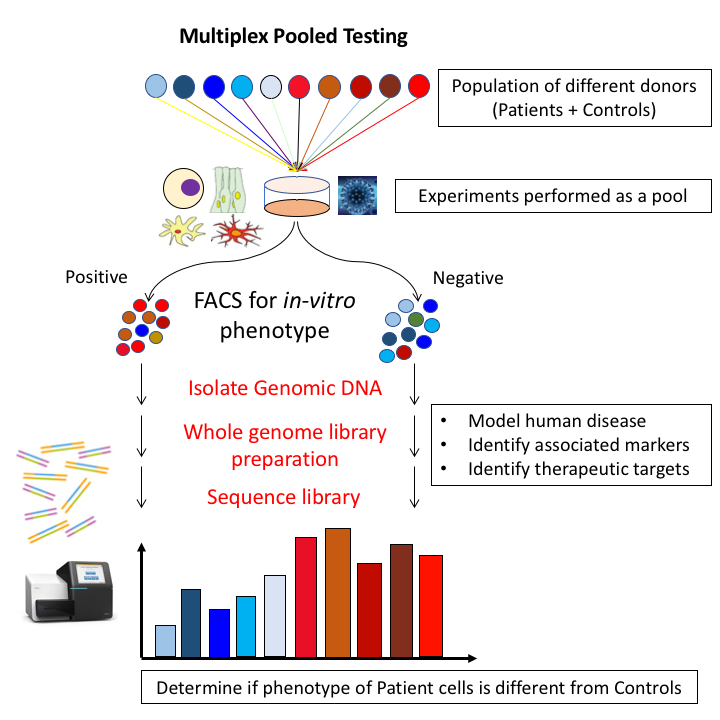
Our lab's research primarily revolves around the use of high-throughput in-vitro modeling of human cell lines for studying human genetic diseases, especially complex diseases. Our goal is to be able to perform multiple in-vitro phenotypic assays on a population of different donor cells to determine if the in-vitro penotype corresponds to the human phenotype. Showing this would be evidence that that the in-vitro model accurately models the human phenotype and further follow up studies using the model would be relevant for studying the human phenotype. To acheive this, our lab currently employs a high-throughput approach where many different donor cells are pooled together for experimentation. The diagram above shows an example where many different patient and control cells are pooled together. The cells are then phenotyped and sorted into Case and Control pools. The genomic DNA from each pool is extracted, library preped and sent for next generation sequencing. Using the resulting sequencing reads, the initial whole-genome sequencing data for each donor individual and a computational algorithm, we can accurately deconvolute donor proportion for each donor within the pool and obtain a phenotypic read-out for each donor. Then, we test whether the patient cells have significantly different phenotype from the control cells (blue versus red). Our current phenotypes include HSV-1 infectivity for predicting Alzheimer's disease as well as Cisplatin induced cell death in otic progenitor cells for predicting Hearing Loss from Cisplatin. The lab is open to exploring more phenotypes and is actively seeking funding and personnel to do so.
Things we do
- Reprogramming cells into induced pluripotent stem cells (iPSCs).
- Differentiating iPSCs into various adult tissue types, e.g. Neurons.
- Phenotypic in-vitro testing of cells, e.g. HSV-1 infection assay.
- Fluorescence-activated cell sorting (FACS) of donor cells into different pools.
- Genomic DNA extraction and library preparation.
- Next-generation sequencing.
- Develop computational methods to analyze sequence data.
- Single-cell RNA sequencing and compuational analysis.
- CRISPR/Cas9 editing of genes to determine effect on phenotype.