Epigenetics and the Non-coding Genome
|
|
|
|
Large-scale segmental genome architecture relates to dark versus light cytogenetic bands and Alu-peaks, implicating both in chromatin regulation. |
|
|
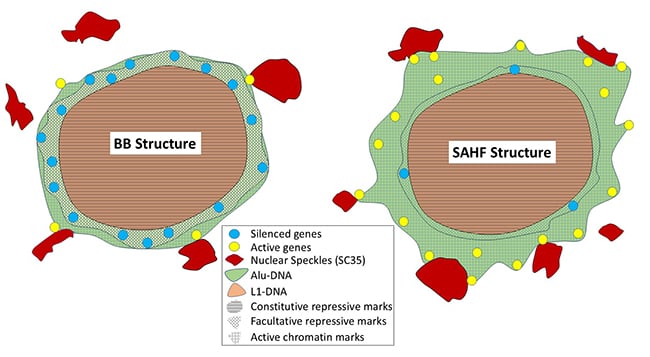
Given Alu and L1 densities differ in cytogenetic bands, we examined their distribution after X-chromosome inactivation or formation of senescent-associated heterochromatin foci (SAHFs). Alu-rich regions remain outside both SAHFs and the Barr Body (BB), affirming that the BB is not the whole chromosome but a condensed, L1-rich core. Hi-C analysis of senescent cells demonstrates large (~10 Mb) G-bands remodel as a contiguous unit, gaining distal intrachromosomal interactions as syntenic G-bands coalesce into SAHFs. Striking peaks of Alu within R-bands strongly resist condensation. |
|
|
Reference: Cytogenetic bands and sharp peaks of Alu underlie large-scale segmental regulation of nuclear genome architecture. Nucleus. 2024 Dec;15(1):2400525. |
|
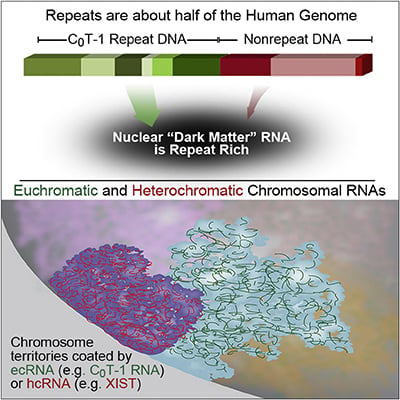
RNA is broadly and stably associated with euchromatin and the predominant component of this chromatin-associated RNA is abundant RNA from interspersed repetitive elements |
|
|
|
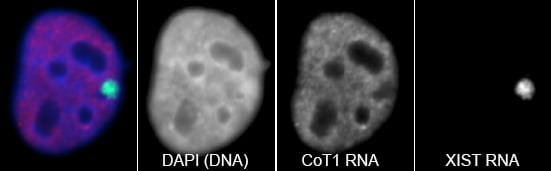
RNA hybridization in situ using CoT-1 (highly repeated) DNA probes detects abundant euchromatin-associated RNA comprised predominantly of repeat sequences (“CoT-1 RNA”), including LINE-1. CoT-1-hybridizing RNA strictly localizes to the interphase chromosome territory in cis, is absent in peripheral heterochromatin and is excluded from chromatin condensed by XIST. Chromosomal RNA associated with euchromatin, comprised largely of CoT-1 repeat RNA, may help maintain open chromatin packaging. |
|
Select references: |
|
Non-coding, repeat-rich sequences in long nascent transcripts play a dynamic structural role in interphase chromosome architecture |
|
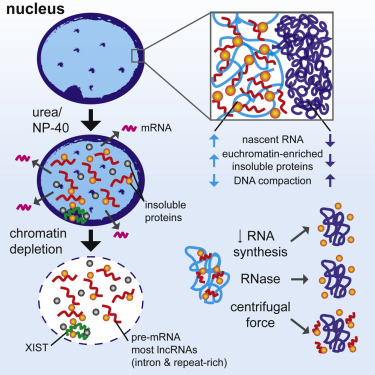
|
We developed a procedure that depletes soluble proteins, chromatin, and most nuclear RNA from the nucleus but does not delocalize XIST, a known architectural RNA, from an insoluble chromosome "scaffold." |
|
Reference: |
|
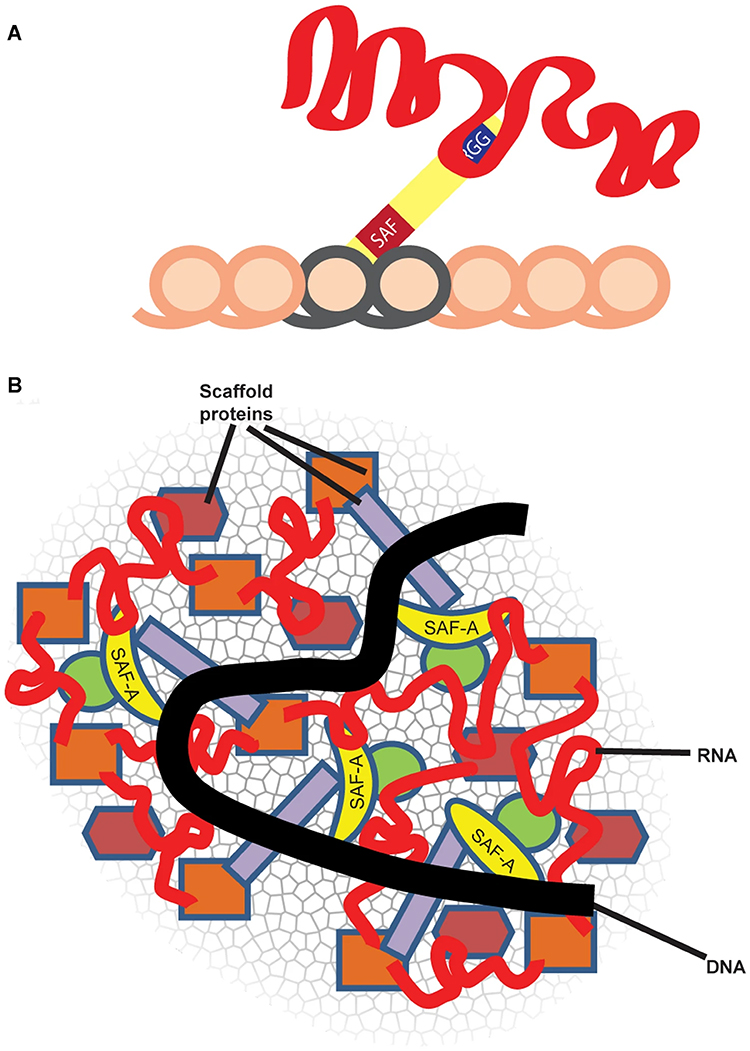
Repeat-rich, chromatin-associated RNAs interact with multiple RNA binding proteins (RBPs) in a complex meshwork integral to chromatin architecture |
|
|
|
SAF-A, also termed heterogenous nuclear protein U (hnRNP U), is a ubiquitous nuclear scaffold protein implicated in XIST RNA localization to the inactive X-chromosome (Xi) but also reported to maintain open DNA packaging in euchromatin. |
|
Reference: |
|
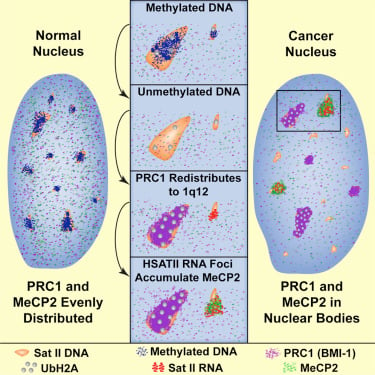
High-copy satellite II (HSATII) sequences bind and impact distribution of chromatin regulatory proteins in cancer |
|
|
We have found that (HSATII) sequences in the human genome can bind and impact distribution of chromatin regulatory proteins and that this goes awry in cancer. In many cancers, master regulatory proteins form two types of cancer-specific nuclear bodies, caused by locus-specific deregulation of HSATII. DNA demethylation at the 1q12 mega-satellite, common in cancer, causes PRC1 aggregation into prominent Cancer-Associated Polycomb (CAP) bodies. These loci remain silent, whereas HSATII loci with reduced PRC1 become derepressed, reflecting imbalanced distribution of UbH2A on these and other PcG-regulated loci. Large nuclear foci of HSATII RNA form and sequester MeCP2 into Cancer-Associated Satellite Transcript (CAST) bodies. Hence, HSATII DNA and RNA have the capacity to act as molecular sponges and sequester chromatin regulatory proteins into abnormal nuclear bodies in cancer.
|
|
|
Reference: |
|
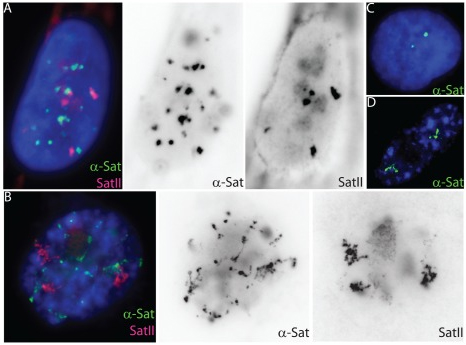
Unfolding of satellite heterochromatin is an early event in cell senescence |
|
|
Cell senescence, the permanent withdrawal of a cell from the cell cycle, is characterized by dramatic, cytological scale changes to DNA condensation throughout the genome, such as the formation of compact Senescent Associated Heterochromatin Foci (SAHF) structures. Our findings showed that SAHF formation is preceded by the unravelling of constitutive heterochromatin into visibly extended structures, which we have termed Senescent Associated Distension of Satellites or SADS. These changes in DNA condensation do not appear to be mediated by changes in heterochromatin-associated histone modifications, but instead may be facilitated by changes in LaminB1 levels and/or other factors that control higher-order chromatin architecture. Cycling (A) and senescent (B) Tig-1 fibroblasts have dramatically different α-satellite (green) and satellite II (red) organization at the resolution of light microscopy. However, closer inspection of gray scale images reveals evidence of organization within each satellite signal, whereby senescent satellites comprise globular domains linked together by threads of more distended DNA. (C–D) Using a probe specific to chromosome 17 α-satellite sequence (green) the signal appears punctate in a cycling cell (C) but distends so that individual domains or globules of still condensed DNA become more visible in senescent cells (D). |
|
|
References: Unfolding the story of chromatin organization in senescent cells. Nucleus. 2015;6(4):254-60. |
|
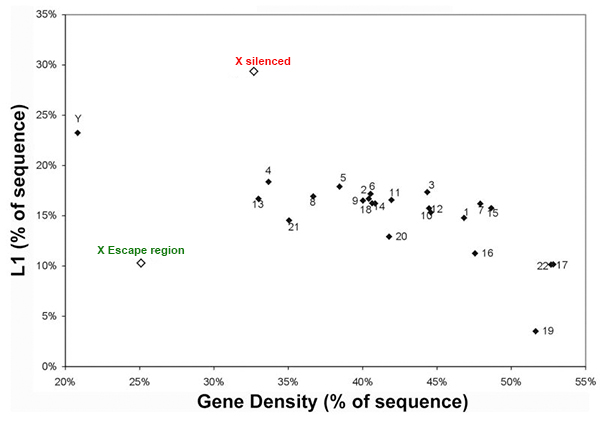
Repeats may facilitate silencing and escape on the X-chromosome |
|
|
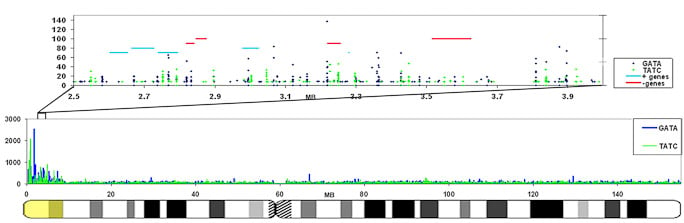
We are interested in studying the potential involvement of specific DNA sequence elements in XIST RNA binding and silencing of the chromosome. A region on the human X wholly escapes inactivation, suggesting sequence specificity in this process. Analysis of all the 9-mer “words” in the human genome suggest LINE and simple sequence repeats are selectively more enriched across the X chromosome. Surprisingly, a singular enrichment of GATA tandem repeats delineate the 8 Mb region that escapes inactivation, suggesting a new paradigm whereby the Xp segment escapes inactivation due to the presence of elements that exclude heterochromatinization, rather than lack of elements that promote it. |
|
|
LINE elements are enriched on the X chromosome except the escape region
|
|
|
The GATA repeat may facilitate escape from silencing within the pseudoautosomal region of the X-chromosome
GATA repeats are 11 fold increased on the region that escapes inactivation compared to the rest of the X-chromosome that is largely silenced. This enrichment is also found in other eutherians that contain a pseudoautosomal region on their X-chromosome. |
|
|
References: |
|