Virus Assembly
Viral Genome Packaging Motors
We have recently initiated studies of machines that ‘walk’ along DNA. These machines generally use ATP to move along DNA and are involved in DNA replication, repair and recombination, as well as viral maturation. Thus, these assemblies are critical for human health. The molecular mechanisms underlying DNA translocation are still mysterious.
The Kelch Lab has developed a novel model system for investigating motor action using a thermophilic virus: phage P74-26. Our recent work has uncovered a trans-activation mechanism for coordinating ATP hydrolysis within the motor ring and has led to proposed mechanisms of DNA translocation and regulation of motor nuclease activity (see Hilbert, Hayes et al PNAS 2015, Hilbert et al Nucl. Acids Res. 2017 and Hayes et al JBC 2020).
Here is a very cool movie from the Bustamante Lab (HHMI/UC Berkeley) depicting how a viral DNA translocase packages DNA into the viral protein shell:
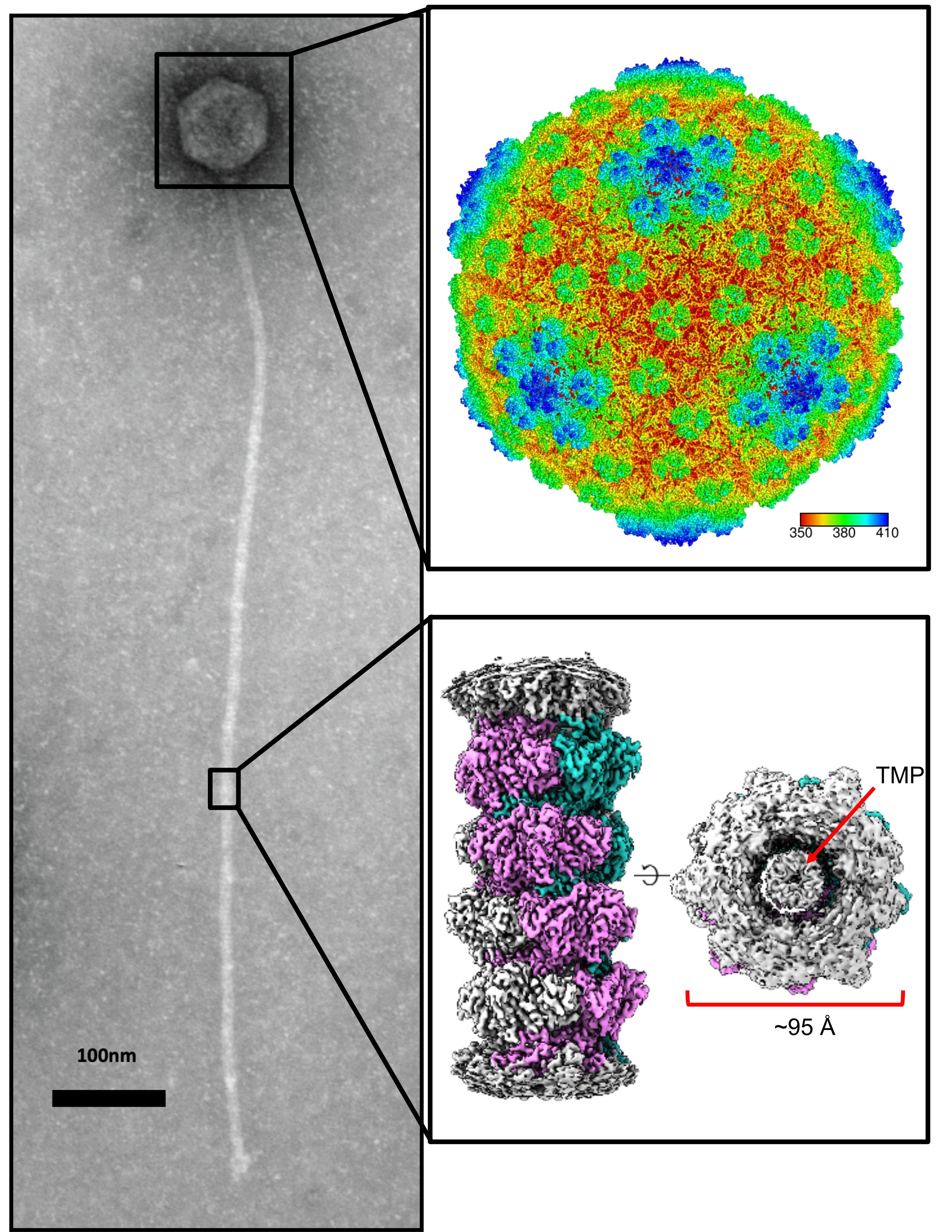
Self-assembly of viral particles
Most known bacteriophage are dsDNA viruses that have protein-shelled capsid that holds the viral genome, and a tail that acts as a conduit to inject that DNA into the host cell. We are interested in the mechanism and properties of these self-assembling systems. Recently, we have investigated self-assembly using the thermophilic phage P74-26, as it is the most thermo-stable known phage. We have determined that the capsid uses an interweaving network of interactions to create an ultra-stable shell (Stone et al Structure 2018 & Stone et al Nat Comm. 2019). We also have taken advantage of the long tail of P74-26 (at nearly 1µm, it is the longest known phage) to investigate the mechanism of tail formation. We have derived a simple structural model that describes a series of auto-inhibitory interactions that regulate the nucleation of tail tubes (Agnello et al JBC 2023; see story in ASBMB Today here). We have also determined the structure of the head-to-tail connection (aka the Neck), which revealed a mechanism for phage to sense pressure during the switch from genome packaging to tail attachment (Sedivy et al biorxiv 2025).