Rules of Gene Expression modeled on human dendritic cell response to pathogens
The developmental shifts that occur when cells respond to environmental stimuli are controlled in large part by gene expression programs involving thousands of genes. Transcription factors (TFs), chromatin modifying enzymes, and cis-acting DNA elements contribute to the networks that underlie such programs. The code that links these variables in such a way that the expression of a given gene can be predicted based on the presence of specific components has yet to be deciphered. A model for such a code will be constructed here based on genome-wide analysis of human dendritic cells (DCs) as they mature in response to pathogens. DCs are antigen-presenting cells that initiate and determine the quality and magnitude of the host immune response. Recent technical advances in stem cell biology, reverse-genetic tools for primary human cells, and genome-wide assessment of transcripts, local chromatin features and long-range chromatin interactions, will be exploited here to construct a model for the transcriptional regulatory network that underlies pathogen detection and maturation in human DCs.
Aim 1
Define the regulatory landscape of hDCs
We will build a comprehensive map of the temporal regulatory landscape of DCs using both chromatin data (ChIP-Seq), three dimensional conformation data (HiC) and expression (RNA-Seq).
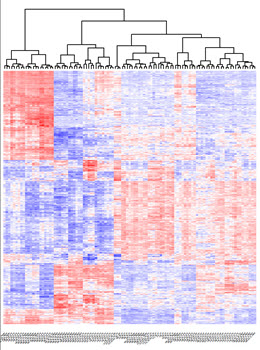
Aim 2
Generate a validated model of trans regulatory factors that drive the differentiation of stimulated hDCs
Using a perturbation approach we will dissect the effect and contribution of trans acting factors to the gene expression network that results when DCs are stimulated by pathogen components.
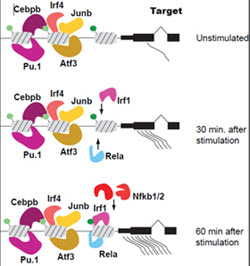
Aim 3
A subset of cis-acting regulatory regions important for controlling the hDC transcriptional response to LPS will be identified using features collected in Aims 1 and 2
We will conduct a targeted, loss of function approach to dissect the contribution of different cis elements to the expression pattern of its targets.
UMass Chan Receives $6.1M

Manuel Garber, Phd and Jeremy Luban, MD
Predicting gene expression in human dendritic cells