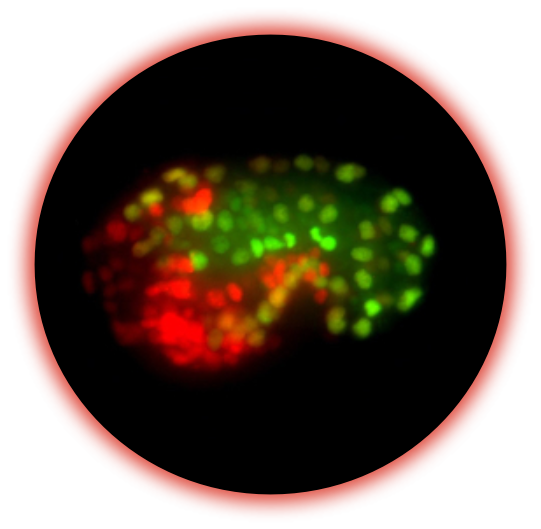
Our long-term goal is to understand how maternal proteins and mRNAs control cell fate in the embryo.
x
Research Areas
Techniques
Genome editing methods including CRISPR/Cas9, MosSCI transgenesis, RNA interference, quantitative phenotyping of worm anatomy and reproductive potential, live imaging of fluorescent reporters, cell fate analysis, immunofluorescence, RNA in situ hybridization, qRT-PCR, protein purification, F-EMSA, fluorescence polarization assays, RNA-seq, polyA-tail sequencing, bioinformatics, and new methods development.