Overview
DNA Replication Kinetics
Eukaryotic genomes replicate in defined patterns, with some parts of the genome replicating early in S phase and other parts replicating later. Replication timing correlates with transcription, chromatin modification, sub-nuclear localization and genome evolution, suggesting an intimate association between replication timing and other important aspects of chromosome metabolism. However, the mechanisms that regulate replication timing are currently poorly understood. We are interested in the biochemical mechanisms that regulate the timing of origin firing, how such mechanisms allow coordination of timing across the genome and how such coordination facilitates robust replication kinetics, ensuring that replication is efficiently completed in all cells.
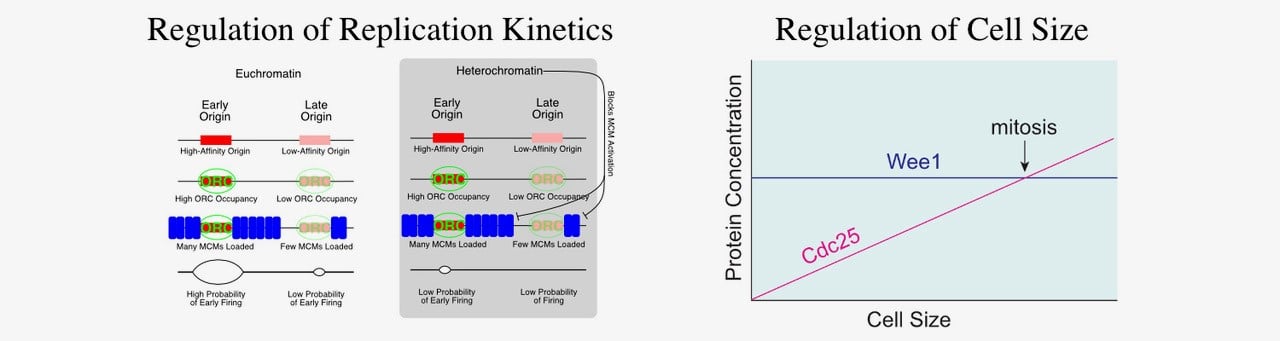
We developed a computational approach to extract replication kinetics from genome-wide replication time courses. Our results support a model where earlier-firing origins have more MCM complexes loaded and a more-accessible chromatin environment. The MCM complex is the replicative helicase; its loading is what establishes sites as potential replication origins. We propose that the timing of origin firing is regulated in by the number of MCM complexes loaded at an origin. Thus, our model suggests a detailed, testable, biochemically plausible mechanism for the regulation of replication timing in eukaryotes. Biochemical validation of our model confirms that early firing origins have more MCM loaded and that changing the number of MCMs loaded at an origin changes its replication timing (Das et al. 2015). We are currently exploring how MCM loading is regulated at different origins.
We are also developing high-throughput, genome-wide, single-molecule approaches to map replication origins and characterize replication kinetics in metazoan genomes (Klein et al. 2017). This new technology will allow test the generality of the mechanisms we discover in yeast to more complicated mammalian genomes.
Cell Size Control
How cells know how big they are is a long-standing biological puzzle. Proper size control is essential for cell viability and cells actively compensate for cell-size perturbations—growing more before division when too small and dividing sooner when to big. Yet, how most cells regulate division in response to size is still a mystery. We have recently shown that, in fission yeast, two direct activators of mitosis, the Cdc13 cyclin and the Cdc25 phosphatase, are expressed in a size-dependent manner (Keifenheim et al. 2017). This manner of expression is unusual because most proteins maintain a constant concentration as cells grow. However, this unusual behavior of two key cell cycle regulators supports the accumulating-activator hypothesis of cell size control. The accumulating-activator hypothesis posits that size-dependent accumulation of a limiting mitotic activator regulates cell size by restraining mitosis when cells are small and express low amounts of the activator, but driving mitosis when cells are large and contain a high amount of the activator. We are testing the hypothesis that Cdc13 and Cdc25 are redundant accumulating activators in fission yeast. We are also investigating how Cdc13 and Cdc25 are expressed in a size dependent manner.