X-ray diffraction of muscle
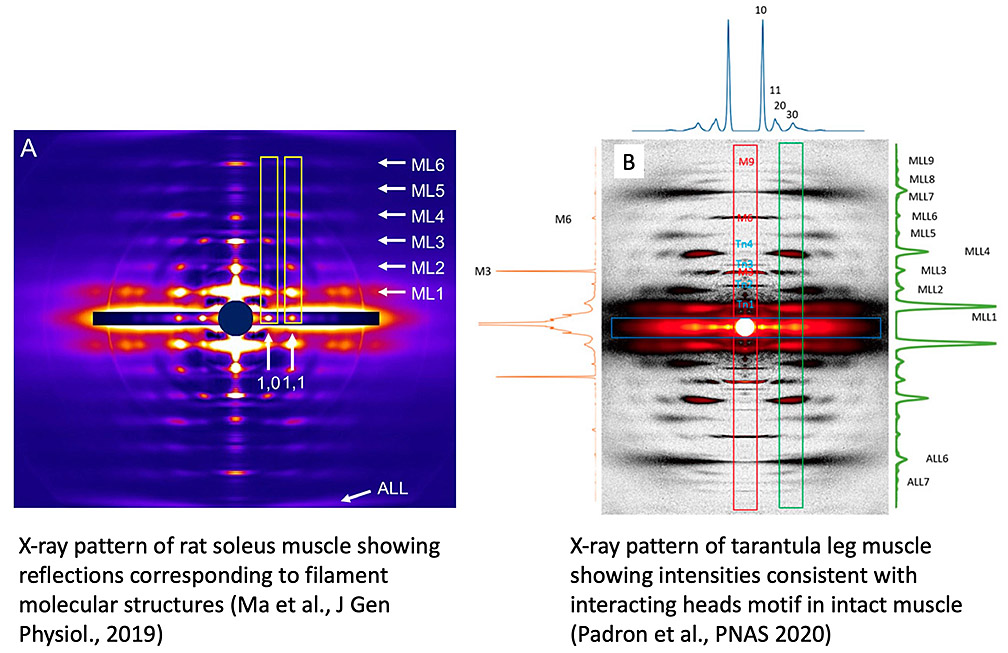
While EM provides the 3D structures of isolated filaments and molecules at high resolution, there could be changes during the isolation procedure. X-ray diffraction yields information on filament structure in the intact muscle and changes in structure when a muscle contracts. The caveat is that filament structure is not directly determined, and depends on model building. We study muscle by high-intensity X-ray diffraction at the Advanced Photon Source (Argonne National Laboratory), in collaboration with Drs. Tom Irving and Weikang Ma. In one study we showed that the thick filaments in mammalian muscle are arranged in specific orientations depending on muscle type (fast or slow; A, below). This was a surprise, with implications for a deeper understanding of the structural basis of muscle physiology (Ma et al, J Gen Physiol, 2019). In another study we modeled the X-ray pattern of tarantula muscle (B, below) to demonstrate the presence of the interacting-heads motif in thick filaments of intact, living muscle (Padron et al., PNAS 2020). This is a crucial finding because the presence of this motif in muscle has recently been challenged, despite its ubiquitous presence in isolated filaments. We showed that this challenge assumed that the myosin heads are much closer to the filament surface than they are in vivo: when this is taken into account, the organization of myosin in IHMs provides excellent agreement with the muscle X-ray pattern (Koubassova et al., Biophys J 2022),. We have now used the new cryo-EM filament model of the human cardiac thick filament to compute the contributions of myosin heads, tails, titin, and cMyBP-C to the diffraction pattern, by including/excluding these components in the calculations. Our results provide a new and objective baseline for understanding the X-ray diffraction pattern of vertebrate muscle (Koubassova, N. A. et al. (2025). Biophys J, doi:10.1016/j.bpj.2025.09.019).