Host-Pathogen Interactions
|
|
Immune recognition of mycobacteria.Both the unique architecture of the mycobacterial cell and its distinct interactions with host phagocytes determine the immune surveillance pathways that are triggered upon infection. We are interested in understanding both the molecular mechanisms underlying this recognition and how the subsequent immune activation influences disease progression.
Mishra, et al. Nitric oxide prevents a pathogen permissive granulocytic inflammation during tuberculosis. Nat Microbiol. 2017 May 15;2:17072 Mishra et al. Nitric oxide controls tuberculosis immunopathology by inhibiting NLRP3 inflammasome-dependent IL-1b processing. Nat Immunol. (2012) Pandey et al. NOD2, RIP2 and IRF5 Play a Critical Role in the Type I Interferon Response to Mycobacterium tuberculosis. PLoS Pathogens. 5(7): e1000500. doi:10.1371/journal.ppat.1000500 (2009). |
|
|
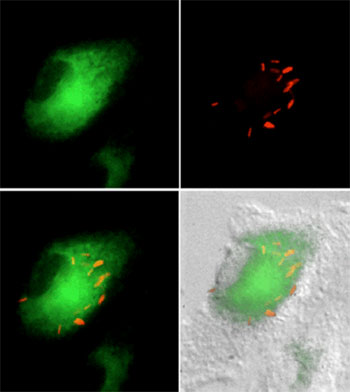 M. tuberculosis (red) is found in cholesterol-rich regions of the macrophage (green) during intracellular growth. |
Nutrient acquisition and carbon metabolism in vivo.Like many other pathogens, M. tuberculosis replicates within an endosome-derived compartment of host cells. A fundamental challenge faced by all of these pathogens is the acquisition of nutrients while surrounded by a host cell membrane. We are investigating the mechanisms used by M. tuberculosis to scavenge nutrients in this niche and to understand how the unusual mixture of available carbon sources alters bacterial metabolism in vivo. Pandey et al. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Nat. Acad. Sci. 105 (11):4376-4380. (2008) Griffin et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathogens. 7(9): e1002251 (2011) Griffin et al. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chemistry and Biology. 19(2):218-27 (2012). Chemistry and Biology (2012) |