Research
Genetic foundations of health and disease
The impacts of most mutations on health and disease are unknown. This hinders our ability to interpret individual genomes, each of which contain hundreds of rare mutations. In the Bolon Lab, we are investigating the biophysical and biochemical underpinnings of gene function. Our studies aim to understand how changes in a protein sequence affect its own function as well as cellular properties and organism health.
Meet the Lab
EMPIRIC approach to measure protein fitness landscapes
A key tool in understanding mutational effects is the ability to generate and analyze mutations. The advent of site-directed mutagenesis methods about thirty years ago ushered in a wave of research associating mutations with effects. The patterns and rules that link mutation to function are complex, which continues to make it challenging to predict the impacts of mutations using computational approaches alone. Experimental analyses continue to be the gold standard for understanding mutational effects. The throughput of traditional one-at-a-time site directed mutation experiments limited the search for the patterns linking mutations and function. To address this critical challenge, we developed the EMPIRIC (Exceedingly Meticulous and Parallel Investigation of Randomized Individual Codons) approach to systematically quantify the effects of all possible individual mutations using a next-generation sequencing readout.
The EMPIRIC approach illuminates local protein fitness landscapes. In the above figure, the effects of each of the 20 possible amino acids as well as stop (nonsense) mutations from an EMPIRIC analyses are presented as a heatmap where yellow indicates high function and blue indicates low function. The inset illustrates the functional selection – in this case the growth rate of cells harboring the mutations.
Relating protein function to cell physiology
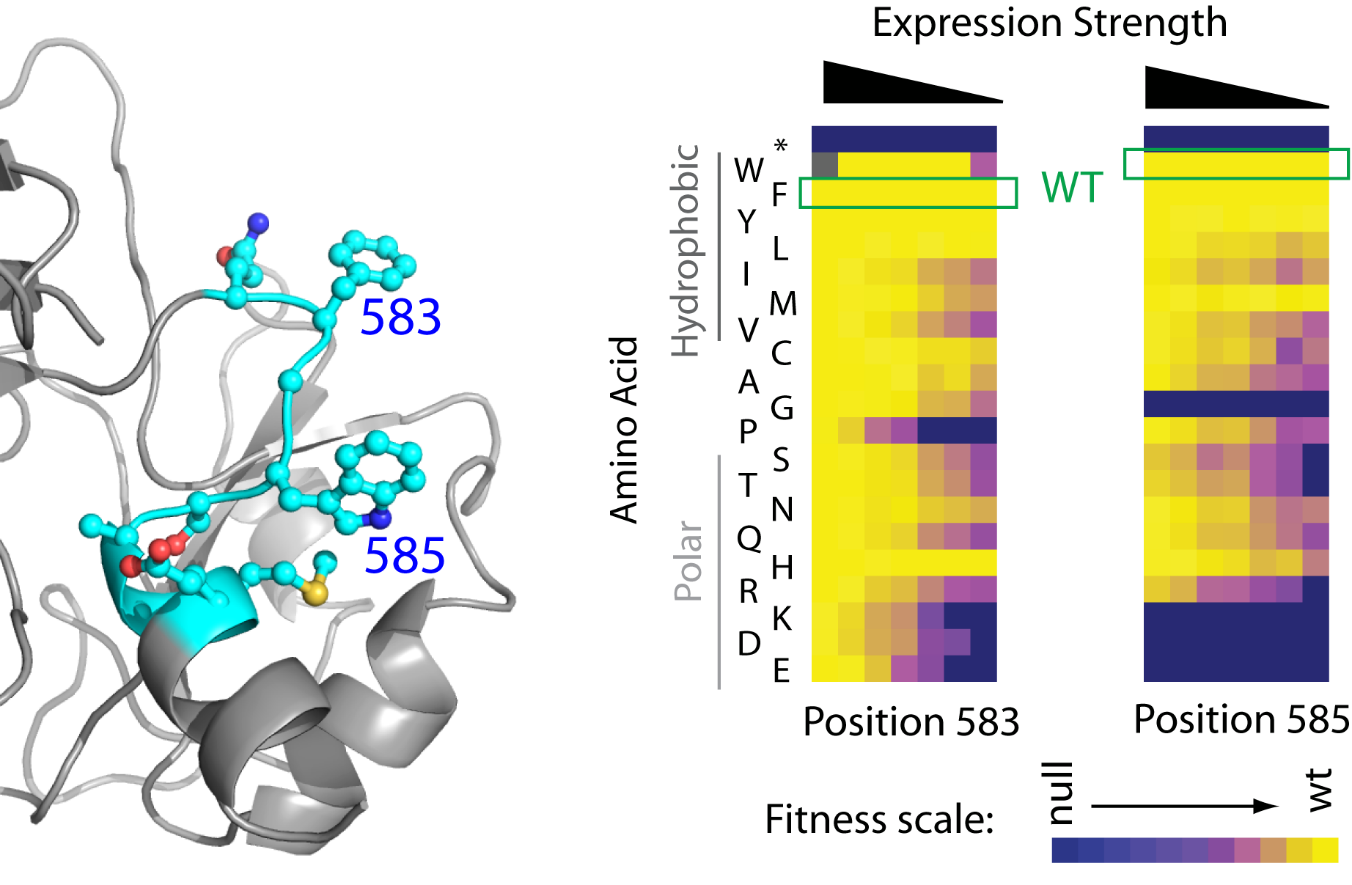
To understand how a specific mutation will impact a cell or an organism, it is not enough to know how the mutation will affect the protein function. We must also know how protein function is linked to cell physiology. Pioneering work by Kacsur and Burns illustrated that for most enzymes in microbes this relationship was far from one to one. We are using the EMPIRIC approach to explore relationships between protein function and cell physiology. In the figure above, we determined how the effects of mutations in Hsp90 on yeast growth were influenced by expression level. At the wild-type expression level (far left column), most amino acids support robust cell growth as illustrated by yellow in the heatmap. At dramatically reduced expression levels, only large hydrophobic amino acids support robust growth at positions 583 and 585. The critical function of these two surface hydrophobic amino acids in binding to hydrophobic patches on substrate proteins is only clear from fitness landscapes under non-standard conditions. We are currently using protein fitness landscapes to explore relationships between protein function, cell physiology, and environmental conditions. We hope that these studies will help reveal the complex biochemical relationships that form the foundation of life.

