HIV Substrate Co-evolution
Interactions between proteins are important for molecular function and hence the binding interfaces between interacting proteins are usually conserved. However, in some instances, mutations in one protein may result in compensatory, correlated mutations in the binding partner and help maintain adequate function.
In HIV-1, the protease binds to and cleaves Gag and Gag-Pol polyproteins at several sites to give rise to functional proteins. However, the protease mutates to evolve resistance to various protease inhibitors. Our work focused on the co-evolution of protease substrate sites within Gag and primary PI-resistance mutations in the protease, and its contribution to PI resistance (Kolli, et.al., 2006, Kolli, et.al., 2009).
Viral sequences derived from infected individuals treated with protease inhibitors were statistically analyzed for co-evolution of mutations in the substrate sites and protease mutations that are the primary cause of PI resistance. Of the sites analyzed, NC-p1 and p1-p6 cleavage site mutations frequently co-evolve with PI resistance protease mutations in different combinations. For example, Leu 449, Ser 451 and/or Pro 453 within p1-p6 cleavage site mutate in a correlated manner with the nelfinavir-resistant D30N/N88D protease.
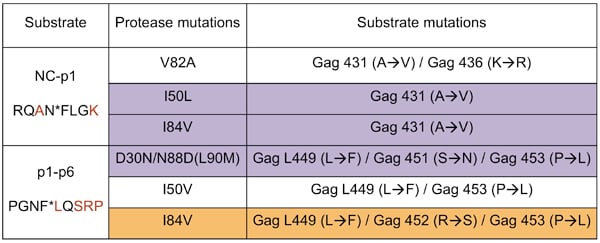
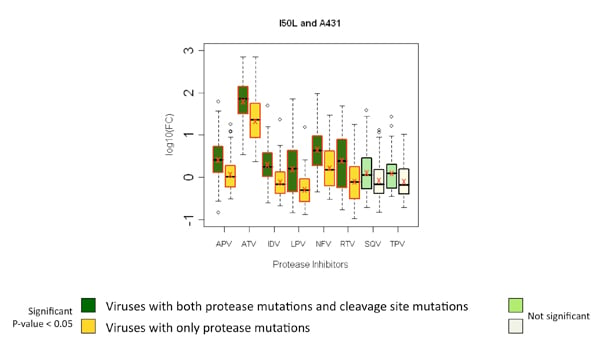
Several of these correlated mutations, when occurring together, were observed to significantly affect phenotypic susceptibility to various PIs. In most instances a significant decrease in phenotypic susceptibility to particular protease inhibitors was observed.
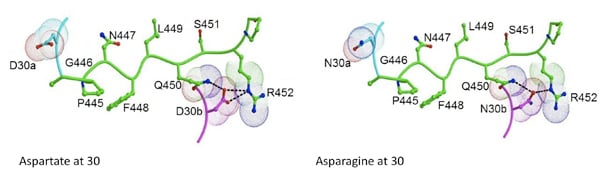
Modeling studies with D30N/N88D protease and the p1-p6 cleavage site suggest that the D30N mutation would likely disrupt the network of hydrogen bonds between Gln 450, Asp 30b and Arg 452. Additionally, L449F and S451N mutations in the p1-p6 cleavage site likely increase the overall interactions with the mutant protease. Currently, structural studies are underway, to probe the underlying structural changes occurring as a result of this co-evolution. Additionally, alteration in Gag processing as a result of this co-evolution is also being investigated.
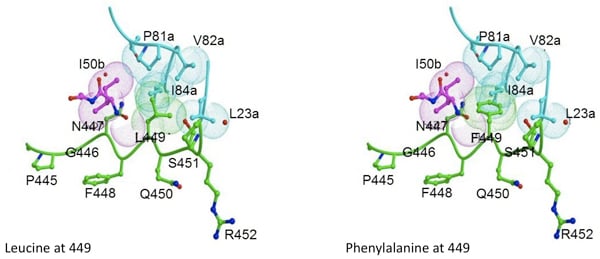
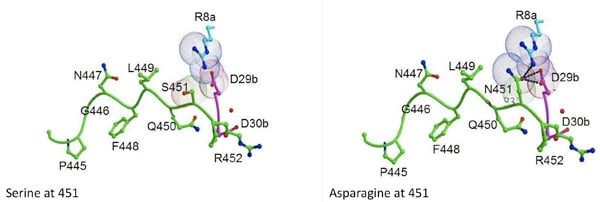
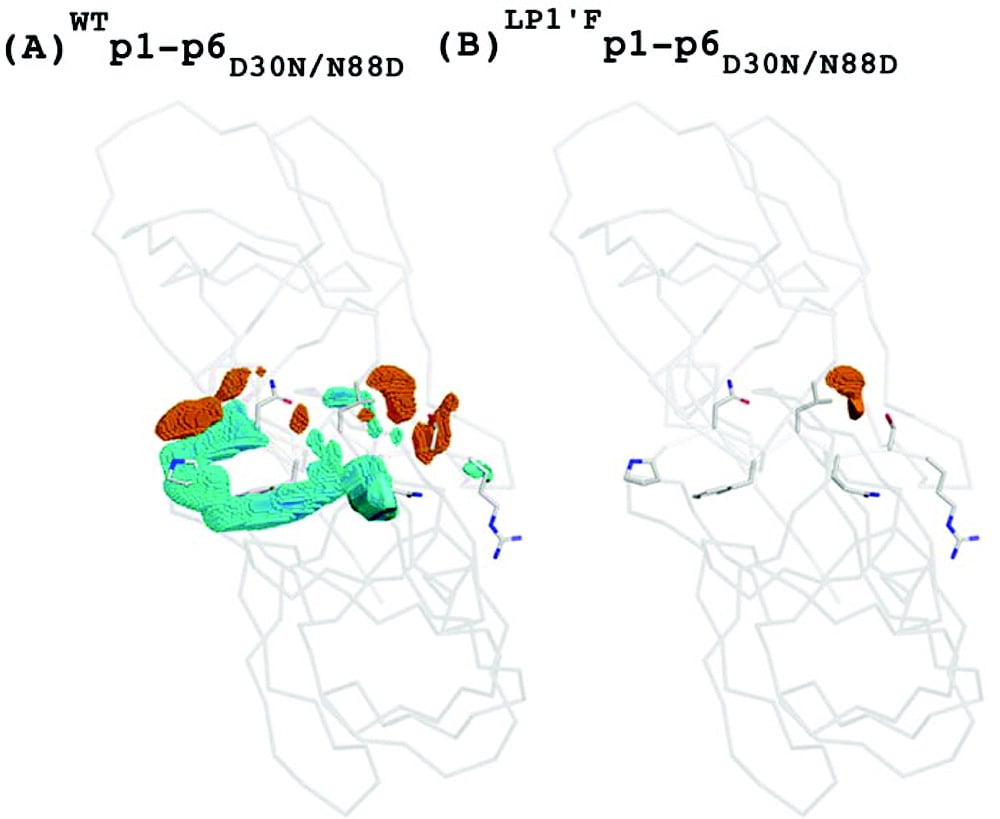
Substrate Co-evolution Validates the Dynamic Substrate Envelope as the Recognition Motif for HIV-1 Protease
The substrate envelope was recently reevaluated by taking the substrate dynamics into account, and the dynamic substrate envelope was reported to better define the substrate specificity for HIV-1 protease (Ozen et al., 2011). Drug resistance occurs mostly through mutations in the protease, occasionally accompanied by cleavage site mutations. Investigation of structural and dynamic properties of three coevolved protease–substrate complexes (AP2VNC-p1V82A, LP1′Fp1-p6D30N/N88D, and SP3′Np1-p6D30N/N88D) show the dynamic substrate envelope is preserved by these cleavage site mutations in the presence of drug-resistance mutations in the protease, if not enhanced. This study on the conformational and mutational ensembles of protease–substrate complexes validates the substrate envelope as the substrate recognition motif for HIV-1 protease. The substrate envelope hypothesis allows for the elucidation of possible drug resistance mutation patterns in the polyprotein cleavage sites (Ozen et al., 2012).