RNA Processing and Ribonucleoprotein Complexes
Research Abstract Summary
Melissa Moore is broadly interested in post-transcriptional gene regulation in eukaryotes via mechanisms involving RNA and RNA-protein (RNP) complexes.
Research Abstract
Our research centers on pre-mRNA processing and large RNA-protein (RNP) complexes. We study their basic structures and functions as well as their contributions to human disease. Current areas of investigation include: (1) single molecule analysis of spliceosome assembly; (2) messenger RNP (mRNP) structure and function; (3) RNP egress by nuclear envelope budding; and (4) development of novel therapeutic approaches targeting RNA-based processes. Enabling these studies, our research spans the disciplines of cell and molecular biology, biochemistry, chemical biology, biophysics and bioinformatics.
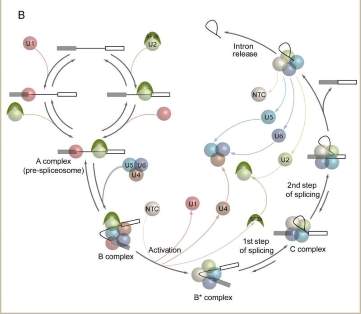
Single Molecule Analysis of Spliceosome Assembly
The majority of protein-coding genes in multicellular organisms contain introns, non-coding regions that must be excised from newly made transcripts (pre-mRNAs) to generate messenger RNAs (mRNAs). Intron excision is mediated by the spliceosome, a highly dynamic macromolecular machine containing five stable small nuclear RNAs (snRNAs) and scores of proteins. By altering the splice sites it utilizes, the spliceosome can produce many different mRNAs from a single gene. Such alternative splicing greatly enhances the number of different proteins that can be encoded within a single genome, and is crucial for development, maintenance and function of diverse tissue types.
A central goal of our research is to elucidate the basic mechanisms by which spliceosomes accurately identify pre-mRNA splice sites and then catalyze intron excision. Recently, we have developed methodologies for fluorescently labeling individual proteins in crude cell lysates. In collaboration with Jeff Gelles (Brandeis), we then use these lysates to monitor spliceosome assembly and intron excision on individual pre-mRNA molecules using multi-wavelength fluorescence Colocalization of Single Molecules Spectroscopy (CoSMoS). By allowing us to analyze the dynamic characteristics of individual spliceosomes in real time, CoSMoS is opening exciting new windows onto previously unaddressable questions regarding spliceosome assembly and internal structural transitions. (This work is supported by a grant from the National Institutes of Health).
mRNP Structure and Function
In addition to removing introns, the process of pre-mRNA splicing loads the mRNA with proteins that fine tune later steps in gene expression. One example is the exon junction complex (EJC), a set of proteins stably deposited by the spliceosome ~24 nucleotides upstream of mRNA exon-exon junctions. By combining RNA immunoprecipitation (RIP) with next generation deep sequencing techniques, we recently mapped EJC deposition sites genome-wide. We find that EJC’s reside at ~80% of exon-exon junctions, as well as at sites that contain sequence motifs targeted by SR proteins, another set of mRNP proteins linked to pre-mRNA splicing. Detailed proteomics reveals that endogenous EJCs interact stably with numerous SR proteins to form multi-megadalton complexes that protect long stretches of spliced mRNAs from nuclease digestion. We are currently investigating the structural nature and functional consequences of these interactions.
RNP Egress by Nuclear Envelope Budding
The canonical view of nucleocytoplasmic transport posits that the sole gateway into and out of the nucleus is the nuclear pore complex (NPC). Thus all RNPs synthesized and assembled in the nucleus have been thought to enter the cytoplasm by transiting the NPC. However, in a collaboration with Vivian Budnik, we recently reported that a subset of endogenous RNPs are able to escape the nucleus by budding through the inner and outer nuclear membranes in a mechanism akin to that used by herpes type viruses. Studies currently underway are designed to identify the complete set of RNAs that utilize the budding pathway and their associated proteins and determine the relationship of nuclear envelope budding to known NPC-dependent export pathways.
RNA and Disease
Aberrant RNAs and RNA binding proteins are important contributors to human disease. Several projects currently underway in the lab are aimed at developing novel therapeutics targeting aberrant RNAs. In collaboration with Neil Aronin and Matt Disney, we are investigating new methods for the elimination of RNAs containing microsatellite expansions. One example is htt mRNA in which expanded CAG repeats lead to Huntington's Disease, an incurable neurodegenerative disorder. Another is C9ORF72, in which a GGGGCC expansion leads to amyotrophic lateral sclerosis (ALS). (This work is supported by grants from the Cure Huntington’s Disease Initiative, CHDI; and the ALS Therapy Alliance).
In collaboration with Ananth Karumanchi, we are working to develop novel treatment modalities for preeclampsia. Preeclampsia, characterized by hypertension, edema and proteinurea after the 20th week of pregnacy is a leading cause of premature birth. It complicates up to 10% of all pregnancies worldwide and is conservatively estimated to cause >75,000 maternal and 500,000 infant deaths globally each year (www.preeclampsia.org). The causative agent is sFlt1 protein, a truncated tyrosine kinase receptor generated by intronic polyadenylation of placental Flt1 pre-mRNA. We are working to limit placental sFlt1 production using RNA silencing approaches. (This work is supported by a grant from the Bill and Melinda Gates Foundation).
Key words:
RNA processing
RNA splicing
Spliceosome
Exon junction complex
Ribonucleoprotein
RNP
Preeclampsia
RNA export
